| MeSH term | MeSH ID | Detail |
|---|---|---|
| Bacterial Infections | D001424 | 21 associated lipids |
| Body Weight | D001835 | 333 associated lipids |
| Drug Hypersensitivity | D004342 | 20 associated lipids |
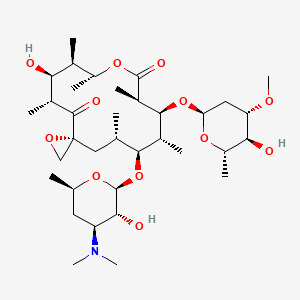
oleandomycin
oleandomycin is a lipid of Polyketides (PK) class. The involved functions are known as 5-(carboxyamino)imidazole ribonucleotide mutase activity, Methylation, enzyme activity, Anabolism and Biosynthetic Pathways. Oleandomycin often locates in Chromosomes and Protoplasm. The associated genes with oleandomycin are resistance genes, Open Reading Frames, CTTNBP2 gene, Gene Feature and Gene Clusters.
Cross Reference
Introduction
To understand associated biological information of oleandomycin, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with oleandomycin?
There are no associated biomedical information in the current reference collection.
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with oleandomycin
PubChem Associated disorders and diseases
What pathways are associated with oleandomycin
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with oleandomycin?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with oleandomycin?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with oleandomycin?
There are no associated biomedical information in the current reference collection.
What genes are associated with oleandomycin?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with oleandomycin?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with oleandomycin
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Tay JH et al. | Regiodivergent Glycosylations of 6-Deoxy-erythronolide B and Oleandomycin-Derived Macrolactones Enabled by Chiral Acid Catalysis. | 2017 | J. Am. Chem. Soc. | pmid:28627172 |
| Bryan MA et al. | Demonstration of non-inferiority of a novel combination intramammary antimicrobial in the treatment of clinical mastitis. | 2016 | N Z Vet J | pmid:27430313 |
| Montemiglio LC et al. | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis. | 2016 | Biochim. Biophys. Acta | pmid:26475642 |
| Bauer J et al. | Impact of stereochemistry on the biological activity of novel oleandomycin derivatives. | 2012 | Bioorg. Med. Chem. | pmid:22377670 |
| Lu S and Zgurskaya HI | Role of ATP binding and hydrolysis in assembly of MacAB-TolC macrolide transporter. | 2012 | Mol. Microbiol. | pmid:23057817 |
| Modali SD and Zgurskaya HI | The periplasmic membrane proximal domain of MacA acts as a switch in stimulation of ATP hydrolysis by MacB transporter. | 2011 | Mol. Microbiol. | pmid:21696464 |
| Berellini G et al. | In silico prediction of volume of distribution in human using linear and nonlinear models on a 669 compound data set. | 2009 | J. Med. Chem. | pmid:19603833 |
| Petrovich YA et al. | Study of drug transport between the blood and lymph in the predominant direction. | 2009 | Bull. Exp. Biol. Med. | pmid:19529861 |
| Shrestha P et al. | Cytochrome P450 (CYP105F2) from Streptomyces peucetius and its activity with oleandomycin. | 2008 | Appl. Microbiol. Biotechnol. | pmid:18437375 |
| Jakeman DL | Sweetly expanding enzymatic glycodiversification. | 2008 | Chem. Biol. | pmid:18420135 |
| Williams GJ et al. | Optimizing glycosyltransferase specificity via "hot spot" saturation mutagenesis presents a catalyst for novobiocin glycorandomization. | 2008 | Chem. Biol. | pmid:18420146 |
| Weidlich M et al. | Differential activity profiles of translation inhibitors in whole-cell and cell-free approaches. | 2008 | Lett. Appl. Microbiol. | pmid:18028330 |
| Ferreira AL et al. | Doxorubicin as an antioxidant: maintenance of myocardial levels of lycopene under doxorubicin treatment. | 2007 | Free Radic. Biol. Med. | pmid:17664138 |
| Williams GJ et al. | Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. | 2007 | Nat. Chem. Biol. | pmid:17828251 |
| González de la Huebra MJ et al. | Sample preparation strategy for the simultaneous determination of macrolide antibiotics in animal feedingstuffs by liquid chromatography with electrochemical detection (HPLC-ECD). | 2007 | J Pharm Biomed Anal | pmid:17257795 |
| Parker KA and Wang P | Deconstruction-reconstruction strategy for accessing valuable polyketides. Preparation of the C15-C24 stereopentad of discodermolide. | 2007 | Org. Lett. | pmid:17924640 |
| Novak P et al. | Systematic approach to understanding macrolide-ribosome interactions: NMR and modeling studies of oleandomycin and its derivatives. | 2006 | J Phys Chem A | pmid:16405329 |
| GarcÃa-Mayor MA et al. | Liquid chromatography-UV diode-array detection method for multi-residue determination of macrolide antibiotics in sheep's milk. | 2006 | J Chromatogr A | pmid:16682049 |
| Novak P et al. | Conformational analysis of oleandomycin and its 8-methylene-9-oxime derivative by NMR and molecular modelling. | 2005 | Org. Biomol. Chem. | pmid:15602597 |
| Kim BS et al. | Identification and antibacterial activity of a new oleandomycin derivative from Streptomyces antibioticus. | 2005 | J. Antibiot. | pmid:15895528 |
| Wang J et al. | Determination of five macrolide antibiotic residues in eggs using liquid chromatography/electrospray ionization tandem mass spectrometry. | 2005 | J. Agric. Food Chem. | pmid:15769104 |
| Yang M et al. | Probing the breadth of macrolide glycosyltransferases: in vitro remodeling of a polyketide antibiotic creates active bacterial uptake and enhances potency. | 2005 | J. Am. Chem. Soc. | pmid:15984838 |
| Tymchyk OV et al. | [Sensitivity of Streptomyces globisporus 3-1--a highly active producer of landomycin E to its own and the other polyketide antibiotics]. | 2004 Mar-Apr | Mikrobiol. Z. | pmid:15208856 |
| Wang J | Determination of five macrolide antibiotic residues in honey by LC-ESI-MS and LC-ESI-MS/MS. | 2004 | J. Agric. Food Chem. | pmid:14733491 |
| Schlüsener MP et al. | Determination of antibiotics from soil by pressurized liquid extraction and liquid chromatography-tandem mass spectrometry. | 2003 | J Chromatogr A | pmid:12899294 |
| González de la Huebra MJ et al. | Comparative study of coulometric and amperometric detection for the determination of macrolides in human urine using high-performance liquid chromatography. | 2003 | Anal Bioanal Chem | pmid:12733015 |
| Long PF et al. | Engineering specificity of starter unit selection by the erythromycin-producing polyketide synthase. | 2002 | Mol. Microbiol. | pmid:11918808 |
| Gaisser S et al. | Parallel pathways for oxidation of 14-membered polyketide macrolactones in Saccharopolyspora erythraea. | 2002 | Mol. Microbiol. | pmid:11994157 |
| Kim YH et al. | Purification and characterization of an erythromycin esterase from an erythromycin-resistant Pseudomonas sp. | 2002 | FEMS Microbiol. Lett. | pmid:12044681 |
| Milanova A and Lashev L | Pharmacokinetics of oleandomycin in dogs after intravenous or oral administration alone and after pretreatment with metamizole or dexamethasone. | 2002 | Vet. Res. Commun. | pmid:11860088 |
| Di Modugno V et al. | Low level resistance to oleandomycin as a marker of ermA in staphylococci. | 2002 | J. Antimicrob. Chemother. | pmid:11815596 |
| Smirnov VV et al. | [Gram-negative eubacteria isolated from the water, molluscs and algae of the Black Sea]. | 2001 Jul-Aug | Mikrobiol. Z. | pmid:11692675 |
| RodrÃguez L et al. | Functional analysis of OleY L-oleandrosyl 3-O-methyltransferase of the oleandomycin biosynthetic pathway in Streptomyces antibioticus. | 2001 | J. Bacteriol. | pmid:11514520 |
| Kimura Y et al. | Characterization of the mac-1 gene encoding a putative ABC transporter from Myxococcus xanthus. | 2001 | J. Biochem. | pmid:11226873 |
| Garzotti M et al. | Coupling of a supercritical fluid chromatography system to a hybrid (Q-TOF 2) mass spectrometer: on-line accurate mass measurements. | 2001 | Rapid Commun. Mass Spectrom. | pmid:11445901 |
| Quirós LM et al. | Glycosylation of macrolide antibiotics. Purification and kinetic studies of a macrolide glycosyltransferase from Streptomyces antibioticus. | 2000 | J. Biol. Chem. | pmid:10766792 |
| Tateda K et al. | Potential of macrolide antibiotics to inhibit protein synthesis of Pseudomonas aeruginosa: suppression of virulence factors and stress response. | 2000 | J. Infect. Chemother. | pmid:11810524 |
| Aguirrezabalaga I et al. | Identification and expression of genes involved in biosynthesis of L-oleandrose and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. | 2000 | Antimicrob. Agents Chemother. | pmid:10770761 |
| Rodriguez L et al. | Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. | 2000 | J. Mol. Microbiol. Biotechnol. | pmid:10937435 |
| Shah S et al. | Cloning, characterization and heterologous expression of a polyketide synthase and P-450 oxidase involved in the biosynthesis of the antibiotic oleandomycin. | 2000 | J. Antibiot. | pmid:10908114 |
| Quirós LM et al. | Inversion of the anomeric configuration of the transferred sugar during inactivation of the macrolide antibiotic oleandomycin catalyzed by a macrolide glycosyltransferase. | 2000 | FEBS Lett. | pmid:10913610 |
| Czeizel AE et al. | A case-control teratological study of spiramycin, roxithromycin, oleandomycin and josamycin. | 2000 | Acta Obstet Gynecol Scand | pmid:10716307 |
| Bajic S et al. | Analysis of erythromycin by liquid chromatography/mass spectrometry using involatile mobile phases with a novel atmospheric pressure ionization source. | 2000 | Rapid Commun. Mass Spectrom. | pmid:10637421 |
| Tang L et al. | Formation of functional heterologous complexes using subunits from the picromycin, erythromycin and oleandomycin polyketide synthases. | 2000 | Chem. Biol. | pmid:10662693 |
| Zhou J et al. | Separation and determination of the macrolide antibiotics (erythromycin, spiramycin and oleandomycin) by capillary electrophoresis coupled with fast reductive voltammetric detection. | 2000 | Electrophoresis | pmid:10826680 |
| Doumith M et al. | Interspecies complementation in Saccharopolyspora erythraea : elucidation of the function of oleP1, oleG1 and oleG2 from the oleandomycin biosynthetic gene cluster of Streptomyces antibioticus and generation of new erythromycin derivatives. | 1999 | Mol. Microbiol. | pmid:10594828 |
| Taniguchi K et al. | Identification of functional amino acids in the macrolide 2'-phosphotransferase II. | 1999 | Antimicrob. Agents Chemother. | pmid:10428938 |
| Taniguchi K et al. | Identification of Escherichia coli clinical isolates producing macrolide 2'-phosphotransferase by a highly sensitive detection method. | 1998 | FEMS Microbiol. Lett. | pmid:9809420 |
| Hoeben D et al. | Antibiotics commonly used to treat mastitis and respiratory burst of bovine polymorphonuclear leukocytes. | 1998 | J. Dairy Sci. | pmid:9532493 |
| Olano C et al. | Analysis of a Streptomyces antibioticus chromosomal region involved in oleandomycin biosynthesis, which encodes two glycosyltransferases responsible for glycosylation of the macrolactone ring. | 1998 | Mol. Gen. Genet. | pmid:9749673 |