| MeSH term | MeSH ID | Detail |
|---|---|---|
| Hemolysis | D006461 | 131 associated lipids |
| Ovarian Neoplasms | D010051 | 10 associated lipids |
| Edema | D004487 | 152 associated lipids |
| Neuroblastoma | D009447 | 66 associated lipids |
| Niemann-Pick Diseases | D009542 | 25 associated lipids |
| Celiac Disease | D002446 | 16 associated lipids |
| Atherosclerosis | D050197 | 85 associated lipids |
| Respiratory Syncytial Virus Infections | D018357 | 10 associated lipids |
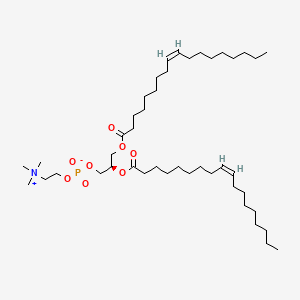
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine is a lipid of Glycerophospholipids (GP) class. 1,2-dioleoyl-sn-glycero-3-phosphocholine is associated with abnormalities such as Exanthema, Renal tubular disorder, Nodule, Gigantism and Mycoses. The involved functions are known as Lysis, Encapsulation, Process, Uptake and Flow or discharge. 1,2-dioleoyl-sn-glycero-3-phosphocholine often locates in Cytoplasmic matrix, Endosomes, soluble, Endoplasmic Reticulum and Membrane. The associated genes with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine are P4HTM gene, synthetic peptide, BCAR1 gene, PCNA gene and CNTNAP1 gene. The related lipids are Liposomes, 1,2-oleoylphosphatidylcholine, 1,2-distearoylphosphatidylethanolamine, Butanols and Cardiolipins. The related experimental models are Mouse Model and Xenograft Model.
Cross Reference
Introduction
To understand associated biological information of 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine is suspected in Gigantism, Nodule, protrusion, DERMATITIS HERPETIFORMIS, FAMILIAL, Exanthema, Renal tubular disorder and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
PubChem Associated disorders and diseases
What pathways are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Xenograft Model
Xenograft Model are used in the study 'Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis.' (Chakravarty D et al., 2011).
Mouse Model
Mouse Model are used in the study 'Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery.' (Landen CN et al., 2005).
Related references are published most in these journals:
| Model | Cross reference | Weighted score | Related literatures |
|---|
NCBI Entrez Crosslinks
All references with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Yamamoto S et al. | Solubilization of Genistein in Phospholipid Vesicles and Their Atioxidant Capacity. | 2019 | J Oleo Sci | pmid:30542013 |
| Vázquez RF et al. | Impact of sphingomyelin acyl chain (16:0 vs 24:1) on the interfacial properties of Langmuir monolayers: A PM-IRRAS study. | 2019 | Colloids Surf B Biointerfaces | pmid:30347381 |
| Becucci L et al. | On the interaction of the highly charged peptides casocidins with biomimetic membranes. | 2018 | Bioelectrochemistry | pmid:29715585 |
| Grasso G et al. | Cell penetrating peptide modulation of membrane biomechanics by Molecular dynamics. | 2018 | J Biomech | pmid:29631749 |
| Ileri Ercan N et al. | Molecular Dynamics Modeling of Methylene Blue-DOPC Lipid Bilayer Interactions. | 2018 | Langmuir | pmid:29553270 |
| Pal S et al. | Effect of Phospholipid Headgroup Charge on the Structure and Dynamics of Water at the Membrane Interface: A Terahertz Spectroscopic Study. | 2018 | J Phys Chem B | pmid:29543460 |
| Donaldson SH and de Aguiar HB | Molecular Imaging of Cholesterol and Lipid Distributions in Model Membranes. | 2018 | J Phys Chem Lett | pmid:29521507 |
| Nugraheni AD et al. | Oxidative modification of methionine80 in cytochrome c by reaction with peroxides. | 2018 | J. Inorg. Biochem. | pmid:29510335 |
| Vafaei S et al. | Hybrid Biomimetic Interfaces Integrating Supported Lipid Bilayers with Decellularized Extracellular Matrix Components. | 2018 | Langmuir | pmid:29489371 |
| Abbott BM et al. | Probing the extended lipid anchorage with cytochrome c and liposomes containing diacylphosphatidylglycerol lipids. | 2018 | Biochim Biophys Acta Biomembr | pmid:29432713 |
| Moghal MMR et al. | Continuous detection of entry of cell-penetrating peptide transportan 10 into single vesicles. | 2018 | Chem. Phys. Lipids | pmid:29425855 |
| Adhyapak PR et al. | Cholesterol induced asymmetry in DOPC bilayers probed by AFM force spectroscopy. | 2018 | Biochim Biophys Acta Biomembr | pmid:29408513 |
| Kohn EM et al. | Role of Cationic Side Chains in the Antimicrobial Activity of C18G. | 2018 | Molecules | pmid:29401708 |
| Bhatia T | An image-processing method to detect sub-optical features based on understanding noise in intensity measurements. | 2018 | Eur. Biophys. J. | pmid:29392337 |
| Bennett M et al. | Molecular clutch drives cell response to surface viscosity. | 2018 | Proc. Natl. Acad. Sci. U.S.A. | pmid:29358406 |
| Cyr N et al. | The hydrophobic region of the peroxin 14: requirements for association with a glycosome mimetic membrane. | 2018 | Biochem. J. | pmid:29259081 |
| Fan X et al. | Ib-AMP4 insertion causes surface rearrangement in the phospholipid bilayer of biomembranes: Implications from quartz-crystal microbalance with dissipation. | 2018 | Biochim Biophys Acta Biomembr | pmid:29106975 |
| Jaschonek S et al. | Intramolecular structural parameters are key modulators of the gel-liquid transition in coarse grained simulations of DPPC and DOPC lipid bilayers. | 2018 | Biochem. Biophys. Res. Commun. | pmid:29101041 |
| Fathizadeh A and Elber R | A mixed alchemical and equilibrium dynamics to simulate heterogeneous dense fluids: Illustrations for Lennard-Jones mixtures and phospholipid membranes. | 2018 | J Chem Phys | pmid:30134684 |
| Meker S et al. | Amyloid-β Peptide Triggers Membrane Remodeling in Supported Lipid Bilayers Depending on Their Hydrophobic Thickness. | 2018 | Langmuir | pmid:30021071 |
| Ichikawa S et al. | Size-dependent uptake of electrically neutral amphipathic polymeric nanoparticles by cell-sized liposomes and an insight into their internalization mechanism in living cells. | 2018 | Chem. Commun. (Camb.) | pmid:29662978 |
| Ting CL et al. | Metastable Prepores in Tension-Free Lipid Bilayers. | 2018 | Phys. Rev. Lett. | pmid:29694074 |
| Sánchez-Murcia PA et al. | Exciton Localization on Ru-Based Photosensitizers Induced by Binding to Lipid Membranes. | 2018 | J Phys Chem Lett | pmid:29363982 |
| Gupta S et al. | Dynamics of Phospholipid Membranes beyond Thermal Undulations. | 2018 | J Phys Chem Lett | pmid:29754484 |
| Halder A et al. | Lipid chain saturation and the cholesterol in the phospholipid membrane affect the spectroscopic properties of lipophilic dye nile red. | 2018 | Spectrochim Acta A Mol Biomol Spectrosc | pmid:28992460 |
| Nagy-Szakal D et al. | Insights into myalgic encephalomyelitis/chronic fatigue syndrome phenotypes through comprehensive metabolomics. | 2018 | Sci Rep | pmid:29968805 |
| Menichetti R et al. | Efficient potential of mean force calculation from multiscale simulations: Solute insertion in a lipid membrane. | 2018 | Biochem. Biophys. Res. Commun. | pmid:28870809 |
| Terakawa MS et al. | Membrane-induced initial structure of α-synuclein control its amyloidogenesis on model membranes. | 2018 | Biochim. Biophys. Acta | pmid:29273335 |
| Bhojoo U et al. | Temperature induced lipid membrane restructuring and changes in nanomechanics. | 2018 | Biochim. Biophys. Acta | pmid:29248477 |
| Barroso MF et al. | Study of lipid peroxidation and ascorbic acid protective role in large unilamellar vesicles from a new electrochemical performance. | 2018 | Bioelectrochemistry | pmid:29247891 |
| Meynaq MYK et al. | Cationic interaction with phosphatidylcholine in a lipid cubic phase studied with electrochemical impedance spectroscopy and small angle X-ray scattering. | 2018 | J Colloid Interface Sci | pmid:29860201 |
| Tesei G et al. | Coarse-grained model of titrating peptides interacting with lipid bilayers. | 2018 | J Chem Phys | pmid:30599743 |
| Lai Y et al. | Highly efficient siRNA transfection in macrophages using apoptotic body-mimic Ca-PS lipopolyplex. | 2018 | Int J Nanomedicine | pmid:30425477 |
| Zhang N et al. | Partition of Glutamic Acid-Based Single-Chain and Gemini Amphiphiles into Phospholipid Membranes. | 2018 | Langmuir | pmid:30350992 |
| Yasuda T et al. | Nanosized Phase Segregation of Sphingomyelin and Dihydrosphigomyelin in Unsaturated Phosphatidylcholine Binary Membranes without Cholesterol. | 2018 | Langmuir | pmid:30350701 |
| Liu X | Interactions of Silver Nanoparticles Formed in Situ on AFM Tips with Supported Lipid Bilayers. | 2018 | Langmuir | pmid:30109936 |
| Melby ES et al. | Peripheral Membrane Proteins Facilitate Nanoparticle Binding at Lipid Bilayer Interfaces. | 2018 | Langmuir | pmid:30102857 |
| Shirane D et al. | Development of an Alcohol Dilution-Lyophilization Method for Preparing Lipid Nanoparticles Containing Encapsulated siRNA. | 2018 | Biol. Pharm. Bull. | pmid:30068880 |
| Maity P et al. | Effect of Zwitterionic Phospholipid on the Interaction of Cationic Membranes with Monovalent Sodium Salts. | 2018 | Langmuir | pmid:30056708 |
| Wang L et al. | Membrane Reconstitution of Monoamine Oxidase Enzymes on Supported Lipid Bilayers. | 2018 | Langmuir | pmid:30049212 |
| Biswas KH et al. | Fabrication of Multicomponent, Spatially Segregated DNA and Protein-Functionalized Supported Membrane Microarray. | 2018 | Langmuir | pmid:30032610 |
| Mobarak E et al. | How to minimize dye-induced perturbations while studying biomembrane structure and dynamics: PEG linkers as a rational alternative. | 2018 | Biochim Biophys Acta Biomembr | pmid:30028957 |
| Wasif Baig M et al. | Orientation of Laurdan in Phospholipid Bilayers Influences Its Fluorescence: Quantum Mechanics and Classical Molecular Dynamics Study. | 2018 | Molecules | pmid:30011800 |
| Da Silveira Cavalcante L et al. | Effect of Liposome Treatment on Hemorheology and Metabolic Profile of Human Red Blood Cells During Hypothermic Storage. | 2018 | Biopreserv Biobank | pmid:30010418 |
| Liu D et al. | Peptide density targets and impedes triple negative breast cancer metastasis. | 2018 | Nat Commun | pmid:29973594 |
| Nomoto T et al. | Effects of Cholesterol Concentration and Osmolarity on the Fluidity and Membrane Tension of Free-standing Black Lipid Membranes. | 2018 | Anal Sci | pmid:29962374 |
| Kluzek M et al. | Kinetic evolution of DOPC lipid bilayers exposed to α-cyclodextrins. | 2018 | Soft Matter | pmid:29947414 |
| Guiselin B et al. | Dynamic Morphologies and Stability of Droplet Interface Bilayers. | 2018 | Phys. Rev. Lett. | pmid:29932701 |
| Bacellar IOL et al. | Permeability of DOPC bilayers under photoinduced oxidation: Sensitivity to photosensitizer. | 2018 | Biochim Biophys Acta Biomembr | pmid:29886032 |
| Mukherjee P et al. | Lipid and membrane recognition by the oxysterol binding protein and its phosphomimetic mutant using dual polarization interferometry. | 2018 | Biochim Biophys Acta Biomembr | pmid:29879417 |