| MeSH term | MeSH ID | Detail |
|---|---|---|
| Hemolysis | D006461 | 131 associated lipids |
| Ovarian Neoplasms | D010051 | 10 associated lipids |
| Edema | D004487 | 152 associated lipids |
| Neuroblastoma | D009447 | 66 associated lipids |
| Niemann-Pick Diseases | D009542 | 25 associated lipids |
| Celiac Disease | D002446 | 16 associated lipids |
| Atherosclerosis | D050197 | 85 associated lipids |
| Respiratory Syncytial Virus Infections | D018357 | 10 associated lipids |
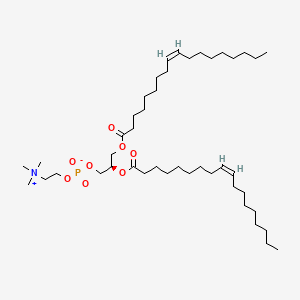
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine is a lipid of Glycerophospholipids (GP) class. 1,2-dioleoyl-sn-glycero-3-phosphocholine is associated with abnormalities such as Exanthema, Renal tubular disorder, Nodule, Gigantism and Mycoses. The involved functions are known as Lysis, Encapsulation, Process, Uptake and Flow or discharge. 1,2-dioleoyl-sn-glycero-3-phosphocholine often locates in Cytoplasmic matrix, Endosomes, soluble, Endoplasmic Reticulum and Membrane. The associated genes with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine are P4HTM gene, synthetic peptide, BCAR1 gene, PCNA gene and CNTNAP1 gene. The related lipids are Liposomes, 1,2-oleoylphosphatidylcholine, 1,2-distearoylphosphatidylethanolamine, Butanols and Cardiolipins. The related experimental models are Mouse Model and Xenograft Model.
Cross Reference
Introduction
To understand associated biological information of 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
1,2-Dioleoyl-sn-Glycero-3-Phosphocholine is suspected in Gigantism, Nodule, protrusion, DERMATITIS HERPETIFORMIS, FAMILIAL, Exanthema, Renal tubular disorder and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
PubChem Associated disorders and diseases
What pathways are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine?
Xenograft Model
Xenograft Model are used in the study 'Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis.' (Chakravarty D et al., 2011).
Mouse Model
Mouse Model are used in the study 'Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery.' (Landen CN et al., 2005).
Related references are published most in these journals:
| Model | Cross reference | Weighted score | Related literatures |
|---|
NCBI Entrez Crosslinks
All references with 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Vafaei S et al. | Hybrid Biomimetic Interfaces Integrating Supported Lipid Bilayers with Decellularized Extracellular Matrix Components. | 2018 | Langmuir | pmid:29489371 |
| Abbott BM et al. | Probing the extended lipid anchorage with cytochrome c and liposomes containing diacylphosphatidylglycerol lipids. | 2018 | Biochim Biophys Acta Biomembr | pmid:29432713 |
| Zhang N et al. | Partition of Glutamic Acid-Based Single-Chain and Gemini Amphiphiles into Phospholipid Membranes. | 2018 | Langmuir | pmid:30350992 |
| Yasuda T et al. | Nanosized Phase Segregation of Sphingomyelin and Dihydrosphigomyelin in Unsaturated Phosphatidylcholine Binary Membranes without Cholesterol. | 2018 | Langmuir | pmid:30350701 |
| Esnault C et al. | Another example of enzymatic promiscuity: the polyphosphate kinase of Streptomyces lividans is endowed with phospholipase D activity. | 2017 | Appl. Microbiol. Biotechnol. | pmid:27488682 |
| Moriwaki H et al. | Electrochemical extraction of gold from wastes as nanoparticles stabilized by phospholipids. | 2017 | Waste Manag | pmid:27424309 |
| Breidigan JM et al. | Influence of the membrane environment on cholesterol transfer. | 2017 | J. Lipid Res. | pmid:29046341 |
| Moleiro LH et al. | Permeability modes in fluctuating lipid membranes with DNA-translocating pores. | 2017 | Adv Colloid Interface Sci | pmid:28735883 |
| Cozza G et al. | Glutathione peroxidase 4-catalyzed reduction of lipid hydroperoxides in membranes: The polar head of membrane phospholipids binds the enzyme and addresses the fatty acid hydroperoxide group toward the redox center. | 2017 | Free Radic. Biol. Med. | pmid:28709976 |
| Winkler PM et al. | Transient Nanoscopic Phase Separation in Biological Lipid Membranes Resolved by Planar Plasmonic Antennas. | 2017 | ACS Nano | pmid:28696660 |
| Chen Y et al. | Interactions of Phospholipid Vesicles with Cationic and Anionic Oligomeric Surfactants. | 2017 | J Phys Chem B | pmid:28686026 |
| Qi W et al. | Curing the Toxicity of Multi-Walled Carbon Nanotubes through Native Small-molecule Drugs. | 2017 | Sci Rep | pmid:28588210 |
| Ramadurai S et al. | Dynamic studies of the interaction of a pH responsive, amphiphilic polymer with a DOPC lipid membrane. | 2017 | Soft Matter | pmid:28327750 |
| Galimzyanov TR et al. | Line Activity of Ganglioside GM1 Regulates the Raft Size Distribution in a Cholesterol-Dependent Manner. | 2017 | Langmuir | pmid:28324651 |
| Suga K et al. | Preferential Adsorption of l-Histidine onto DOPC/Sphingomyelin/3β-[N-(N',N'-dimethylaminoethane)carbamoyl]cholesterol Liposomes in the Presence of Chiral Organic Acids. | 2017 | Langmuir | pmid:28272888 |
| Osella S and Knippenberg S | Triggering On/Off States of Photoswitchable Probes in Biological Environments. | 2017 | J. Am. Chem. Soc. | pmid:28252300 |
| Lütgebaucks C et al. | Characterization of the interface of binary mixed DOPC:DOPS liposomes in water: The impact of charge condensation. | 2017 | J Chem Phys | pmid:28147550 |
| Potvin-Fournier K et al. | Membrane fluidity is a driving force for recoverin myristoyl immobilization in zwitterionic lipids. | 2017 | Biochem. Biophys. Res. Commun. | pmid:28684313 |
| Avakyan N et al. | Long-Range Ordering of Blunt-Ended DNA Tiles on Supported Lipid Bilayers. | 2017 | J. Am. Chem. Soc. | pmid:28783358 |
| Larsen JB et al. | Membrane Curvature and Lipid Composition Synergize To Regulate N-Ras Anchor Recruitment. | 2017 | Biophys. J. | pmid:28738989 |
| Korshavn KJ et al. | Reduced Lipid Bilayer Thickness Regulates the Aggregation and Cytotoxicity of Amyloid-β. | 2017 | J. Biol. Chem. | pmid:28154182 |
| Zhang X et al. | Islet Amyloid Polypeptide Membrane Interactions: Effects of Membrane Composition. | 2017 | Biochemistry | pmid:28054763 |
| Rudolphi-Skórska E et al. | The Effects of the Structure and Composition of the Hydrophobic Parts of Phosphatidylcholine-Containing Systems on Phosphatidylcholine Oxidation by Ozone. | 2017 | J. Membr. Biol. | pmid:28799139 |
| McDonald C et al. | Structure and function of PspA and Vipp1 N-terminal peptides: Insights into the membrane stress sensing and mitigation. | 2017 | Biochim. Biophys. Acta | pmid:27806910 |
| Li H et al. | Drude Polarizable Force Field for Molecular Dynamics Simulations of Saturated and Unsaturated Zwitterionic Lipids. | 2017 | J Chem Theory Comput | pmid:28731702 |
| Wallner J et al. | An approach for liposome immobilization using sterically stabilized micelles (SSMs) as a precursor for bio-layer interferometry-based interaction studies. | 2017 | Colloids Surf B Biointerfaces | pmid:28340485 |
| Wagner MJ et al. | Preclinical Mammalian Safety Studies of EPHARNA (DOPC Nanoliposomal EphA2-Targeted siRNA). | 2017 | Mol. Cancer Ther. | pmid:28265009 |
| Malyshka D and Schweitzer-Stenner R | Ferrocyanide-Mediated Photoreduction of Ferricytochrome C Utilized to Selectively Probe Non-native Conformations Induced by Binding to Cardiolipin-Containing Liposomes. | 2017 | Chemistry | pmid:27859757 |
| Khan FI et al. | Structure prediction and functional analyses of a thermostable lipase obtained from Shewanella putrefaciens. | 2017 | J. Biomol. Struct. Dyn. | pmid:27366981 |
| Krishnan P et al. | Hetero-multivalent binding of cholera toxin subunit B with glycolipid mixtures. | 2017 | Colloids Surf B Biointerfaces | pmid:28946063 |
| Clulow AJ et al. | Characterization of Solubilizing Nanoaggregates Present in Different Versions of Simulated Intestinal Fluid. | 2017 | J Phys Chem B | pmid:29090933 |
| Ding W et al. | Effects of High Pressure on Phospholipid Bilayers. | 2017 | J Phys Chem B | pmid:28926699 |
| Navon Y et al. | pH-Sensitive Interactions between Cellulose Nanocrystals and DOPC Liposomes. | 2017 | Biomacromolecules | pmid:28799758 |
| Xing J et al. | Increasing vaccine production using pulsed ultrasound waves. | 2017 | PLoS ONE | pmid:29176801 |
| Massiot J et al. | Impact of lipid composition and photosensitizer hydrophobicity on the efficiency of light-triggered liposomal release. | 2017 | Phys Chem Chem Phys | pmid:28425533 |
| Lum K et al. | Exchange of Gramicidin between Lipid Bilayers: Implications for the Mechanism of Channel Formation. | 2017 | Biophys. J. | pmid:29045870 |
| He B et al. | Increased cellular uptake of peptide-modified PEGylated gold nanoparticles. | 2017 | Biochem. Biophys. Res. Commun. | pmid:28993197 |
| Cornell CE et al. | n-Alcohol Length Governs Shift in L-L Mixing Temperatures in Synthetic and Cell-Derived Membranes. | 2017 | Biophys. J. | pmid:28801104 |
| Moniruzzaman M et al. | Entry of a Six-Residue Antimicrobial Peptide Derived from Lactoferricin B into Single Vesicles and Escherichia coli Cells without Damaging their Membranes. | 2017 | Biochemistry | pmid:28752991 |
| Lee MT et al. | Molecular State of the Membrane-Active Antibiotic Daptomycin. | 2017 | Biophys. J. | pmid:28700928 |
| Ritzmann N et al. | Fusion Domains Guide the Oriented Insertion of Light-Driven Proton Pumps into Liposomes. | 2017 | Biophys. J. | pmid:28697898 |
| Elderdfi M et al. | MPP1 interacts with DOPC/SM/Cholesterol in an artificial membrane system using Langmuir-Blodgett monolayer. | 2017 | Gen. Physiol. Biophys. | pmid:28653654 |
| Lindberg DJ et al. | Lipid membranes catalyse the fibril formation of the amyloid-β (1-42) peptide through lipid-fibril interactions that reinforce secondary pathways. | 2017 | Biochim. Biophys. Acta | pmid:28564579 |
| Przybyło M et al. | Changes in lipid membrane mechanics induced by di- and tri-phenyltins. | 2017 | Biochim. Biophys. Acta | pmid:28461050 |
| HÄ…c-Wydro K et al. | Effect of Cd and Cd/auxin mixtures on lipid monolayers - Model membrane studies on the role of auxins in phytoremediation of metal ions from contaminated environment. | 2017 | Biochim. Biophys. Acta | pmid:28343956 |
| Alvares DS et al. | Phosphatidylserine lipids and membrane order precisely regulate the activity of Polybia-MP1 peptide. | 2017 | Biochim. Biophys. Acta | pmid:28274844 |
| Korytowski A et al. | Accumulation of phosphatidylcholine on gut mucosal surface is not dominated by electrostatic interactions. | 2017 | Biochim. Biophys. Acta | pmid:28212861 |
| Chen X et al. | Efficient internalization of TAT peptide in zwitterionic DOPC phospholipid membrane revealed by neutron diffraction. | 2017 | Biochim. Biophys. Acta | pmid:28153495 |
| Rashid A et al. | Substituents modulate biphenyl penetration into lipid membranes. | 2017 | Biochim. Biophys. Acta | pmid:28130013 |
| Haney CM et al. | Site-Specific Fluorescence Polarization for Studying the Disaggregation of α-Synuclein Fibrils by Small Molecules. | 2017 | Biochemistry | pmid:28045494 |