| MeSH term | MeSH ID | Detail |
|---|---|---|
| Diabetes Mellitus, Type 2 | D003924 | 87 associated lipids |
| Epilepsy | D004827 | 35 associated lipids |
| Fatty Liver | D005234 | 48 associated lipids |
| Fatty Liver, Alcoholic | D005235 | 11 associated lipids |
| Galactosemias | D005693 | 5 associated lipids |
| Hamartoma Syndrome, Multiple | D006223 | 1 associated lipids |
| Hemolysis | D006461 | 131 associated lipids |
| Lipid Metabolism, Inborn Errors | D008052 | 26 associated lipids |
| Lupus Erythematosus, Systemic | D008180 | 43 associated lipids |
| Mammary Neoplasms, Experimental | D008325 | 67 associated lipids |
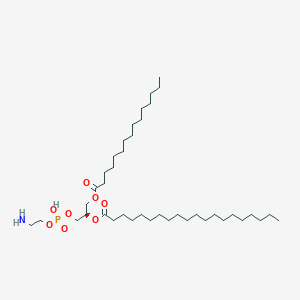
PE(15:0/20:0)
PE(15:0/20:0) is a lipid of Glycerophospholipids (GP) class. Pe(15:0/20:0) is associated with abnormalities such as Exanthema, Infection, Painful Bladder Syndrome, Obesity and Fatty Liver. The involved functions are known as conjugation, Transcription, Genetic, Sinking, Autophagy and Protein Biosynthesis. Pe(15:0/20:0) often locates in membrane fraction, soluble, Membrane, Body tissue and Tissue membrane. The associated genes with PE(15:0/20:0) are GABARAPL2 gene, ATG10 gene, ATG12 gene, SLC33A1 gene and GABARAP gene. The related lipids are Liposomes, Lipopolysaccharides, Phosphatidylserines, Membrane Lipids and Cardiolipins. The related experimental models are Knock-out and Cancer Model.
Cross Reference
Introduction
To understand associated biological information of PE(15:0/20:0), we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with PE(15:0/20:0)?
PE(15:0/20:0) is suspected in Infection, CONE-ROD DYSTROPHY 1 (disorder), Diabetes, Obesity, Malaria, Atherosclerosis and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with PE(15:0/20:0)
PubChem Associated disorders and diseases
What pathways are associated with PE(15:0/20:0)
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with PE(15:0/20:0)?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with PE(15:0/20:0)?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with PE(15:0/20:0)?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with PE(15:0/20:0)?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with PE(15:0/20:0)?
Knock-out
Knock-out are used in the study 'Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo.' (Hörl G et al., 2011) and Knock-out are used in the study 'An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure.' (Fujita N et al., 2008).
Cancer Model
Cancer Model are used in the study 'Improving penetration in tumors with nanoassemblies of phospholipids and doxorubicin.' (Tang N et al., 2007).
Related references are published most in these journals:
| Model | Cross reference | Weighted score | Related literatures |
|---|
NCBI Entrez Crosslinks
All references with PE(15:0/20:0)
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Choi S et al. | Dokdonia aurantiaca sp. nov., isolated from seaweed Zostera marina. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29570445 |
| Sharma A et al. | Subsaxibacter sediminis sp. nov., isolated from Arctic glacial sediment and emended description of the genus Subsaxibacter. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29561259 |
| Zhao JC et al. | Flavobacterium artemisiae sp. nov., isolated from the rhizosphere of Artemisia annua L. and emended descriptions of Flavobacterium compostarboris and Flavobacterium procerum. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29547095 |
| Mioka T et al. | Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. | 2018 | Mol. Biol. Cell | pmid:29540528 |
| Fajardo VA et al. | Elevated whole muscle phosphatidylcholine: phosphatidylethanolamine ratio coincides with reduced SERCA activity in murine overloaded plantaris muscles. | 2018 | Lipids Health Dis | pmid:29534725 |
| Yu CY et al. | Membrane glycerolipid equilibrium under endoplasmic reticulum stress in Arabidopsis thaliana. | 2018 | Biochem. Biophys. Res. Commun. | pmid:29524407 |
| Shin SK et al. | Tenacibaculum todarodis sp. nov., isolated from a squid. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29521615 |
| Park S et al. | Pseudobizionia ponticola gen. nov., sp. nov., isolated from seawater. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29521613 |
| Burger HM et al. | Modulation of key lipid raft constituents in primary rat hepatocytes by fumonisin B - Implications for cancer promotion in the liver. | 2018 | Food Chem. Toxicol. | pmid:29510220 |
| Park S et al. | Thalassotalea insulae sp. nov., isolated from tidal flat sediment. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29488866 |
| Cai H et al. | Flavobacterium cyanobacteriorum sp. nov., isolated from cyanobacterial aggregates in a eutrophic lake. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29485392 |
| Kim HC et al. | Vitellibacter todarodis sp. nov., isolated from intestinal tract of a squid (Todarodes pacificus). | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29480794 |
| Kim YO et al. | Bizionia berychis sp. nov., isolated from intestinal tract of a splendid alfonsino (Beryx splendens). | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29469688 |
| Li W et al. | Spirosoma horti sp. nov., isolated from apple orchard soil. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458662 |
| Cui MD et al. | Pedobacter agrisoli sp. nov., isolated from farmland soil. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458546 |
| Jiang WK et al. | Terrimonas soli sp. nov., isolated from farmland soil. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458527 |
| Choi JY et al. | Flavobacterium kingsejongi sp. nov., a carotenoid-producing species isolated from Antarctic penguin faeces. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458488 |
| Nedashkovskaya OI et al. | Aquimarina algiphila sp. nov., a chitin degrading bacterium isolated from the red alga Tichocarpus crinitus. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458485 |
| Choi S et al. | Dokdonia flava sp. nov., isolated from the seaweed Zostera marina. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458481 |
| Kwon YM et al. | Euzebyella algicola sp. nov., a marine bacterium of the family Flavobacteriaceae, isolated from green algae. | 2018 | Int. J. Syst. Evol. Microbiol. | pmid:29458477 |