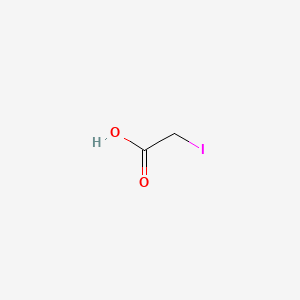
Iodoacetic acid
Iodoacetic acid is a lipid of Fatty Acyls (FA) class. Iodoacetic acid is associated with abnormalities such as Photoreceptor degeneration and Post MI. The involved functions are known as Hypoxia, Glycolysis, Metabolic Inhibition, Oxidation and PTPS activity. Iodoacetic acid often locates in Extracellular, Muscle, Mitochondria, Cytoplasmic matrix and Tissue membrane. The associated genes with Iodoacetic acid are SLC33A1 gene, GTF2I gene, Mutant Proteins, TRIM33 gene and oxytocin, 1-desamino-(O-Et-Tyr)(2)-.
Cross Reference
Introduction
To understand associated biological information of Iodoacetic acid, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with Iodoacetic acid?
Iodoacetic acid is suspected in Photoreceptor degeneration, Post MI and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with Iodoacetic acid
PubChem Associated disorders and diseases
What pathways are associated with Iodoacetic acid
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with Iodoacetic acid?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with Iodoacetic acid?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with Iodoacetic acid?
There are no associated biomedical information in the current reference collection.
What genes are associated with Iodoacetic acid?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with Iodoacetic acid?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with Iodoacetic acid
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Sammut EB and Kannegieter NJ | Use of sodium monoiodoacetate to fuse the distal hock joints in horses. | 1995 | Aust. Vet. J. | pmid:8787522 |
| Whitton C | Chemical arthrodesis of the distal tarsal joints using sodium monoiodoacetate in 104 horses. | 2004 | Aust. Vet. J. | pmid:15181929 |
| Thomas JA and Park EM | Oxy radical-initiated protein S-thiolation and enzymic dethiolation. | 1988 | Basic Life Sci. | pmid:3250494 |
| Lipasti JA et al. | The relationship between ischemic contracture and no-reflow phenomenon in isolated rat heart. | 1982 Jul-Aug | Basic Res. Cardiol. | pmid:7150210 |
| Regling G et al. | [Monoiodoacetic acid-induced arthropathy of the rabbit knee--a contribution to the pathogenesis of arthrosis]. | 1989 | Beitr Orthop Traumatol | pmid:2751625 |
| Laiho KU and Trump BF | Mitochondrial changes, ion and water shifts in the cellular injury of ehrlich ascites tumor cells. | 1975 | Beitr Pathol | pmid:170904 |
| Talwar HS et al. | Sodium glycolate absorption in rat intestine. | 1984 | Biochem Med | pmid:6477536 |
| Chew A et al. | Mutations in a putative zinc-binding domain inactivate the mitochondrial intermediate peptidase. | 1996 | Biochem. Biophys. Res. Commun. | pmid:8831696 |
| Brodie AE and Reed DJ | Reversible oxidation of glyceraldehyde 3-phosphate dehydrogenase thiols in human lung carcinoma cells by hydrogen peroxide. | 1987 | Biochem. Biophys. Res. Commun. | pmid:3675570 |
| Canfield LM | Vitamin K-dependent oxygenase/carboxylase; differential inactivation by sulfhydryl reagents. | 1987 | Biochem. Biophys. Res. Commun. | pmid:3675572 |
| Yasuhara T and Ohashi A | New chelator-sensitive proteases in matrix of yeast mitochondria. | 1987 | Biochem. Biophys. Res. Commun. | pmid:3555485 |
| Villalba JM et al. | Thiol groups are involved in NADH-ascorbate free radical reductase activity of rat liver plasma membrane. | 1993 | Biochem. Biophys. Res. Commun. | pmid:8484777 |
| Ding PZ | An investigation of cysteine mutants on the cytoplasmic loop X/XI in the melibiose transporter of Escherichia coli by using thiol reagents: implication of structural conservation of charged residues. | 2003 | Biochem. Biophys. Res. Commun. | pmid:12878191 |
| Nyberg F et al. | Endopeptidase in human cerebrospinal fluid which cleaves proenkephalin B opioid peptides at consecutive basic amino acids. | 1985 | Biochem. Biophys. Res. Commun. | pmid:2864927 |
| Advani S and Roy KB | Properties and secondary structure analysis of BanI endonuclease: identification of putative active site. | 2000 | Biochem. Biophys. Res. Commun. | pmid:11112410 |
| Haniu M et al. | Heme binding and substrate-protected cysteine residues in P-450cam. | 1983 | Biochem. Biophys. Res. Commun. | pmid:6639664 |
| Fisher JM et al. | Evidence for a membrane sulfhydryl associated with resistance to melphalan in a murine L1210 leukemia line. | 1983 | Biochem. Biophys. Res. Commun. | pmid:6667269 |
| Ikeda U et al. | Identification of two new calcium dependent hydrolases in the human heart. | 1986 | Biochem. Biophys. Res. Commun. | pmid:3010997 |
| Pasero L et al. | Localization of the two free thiol groups in the porcine pancreatic alpha-amylase I sequence. | 1983 | Biochem. Biophys. Res. Commun. | pmid:6188459 |
| Tomlinson G and Kinsch EM | S-mercuric-N-dansyl-cysteine labels the free sulfhydryl groups of human serum cholinesterase. | 1989 | Biochem. Biophys. Res. Commun. | pmid:2916998 |
| Harrison DC et al. | A pH-dependent phospholipase A2 contributes to loss of plasma membrane integrity during chemical hypoxia in rat hepatocytes. | 1991 | Biochem. Biophys. Res. Commun. | pmid:1899571 |
| Jha S et al. | Covalent modification of cysteine 193 impairs ATPase function of nucleotide-binding domain of a Candida drug efflux pump. | 2003 | Biochem. Biophys. Res. Commun. | pmid:14550284 |
| Seya T and Nagasawa S | Isolation of an iC3b forming enzyme from peritoneal polymorphonuclear leukocytes of guinea pigs. | 1983 | Biochem. Biophys. Res. Commun. | pmid:6229252 |
| Güther T et al. | Regulation of intracellular magnesium by Mg2+ efflux. | 1984 | Biochem. Biophys. Res. Commun. | pmid:6422934 |
| Kuzminskaya EV et al. | Rabbit muscle D-glyceraldehyde-3-phosphate dehydrogenase: half-of-the-sites reactivity of the enzyme modified at arginine residues. | 1992 | Biochem. Biophys. Res. Commun. | pmid:1530616 |
| Kirley JW and Day RA | Irreversible inhibition of carbonic anhydrase by the carbon dioxide analog cyanogen. | 1985 | Biochem. Biophys. Res. Commun. | pmid:3918532 |
| Holtzer ME et al. | The effect of sulfhydryl blocking groups on the thermal unfolding of alpha alpha tropomyosin coiled coils. | 1990 | Biochem. Biophys. Res. Commun. | pmid:2306243 |
| Kyprianou N and Isaacs JT | "Thymineless" death in androgen-independent prostatic cancer cells. | 1989 | Biochem. Biophys. Res. Commun. | pmid:2531584 |
| Harrsch PB et al. | Amino acid sequence similarity between spinach chloroplast and mammalian gluconeogenic fructose-1,6-bisphosphatase. | 1985 | Biochem. Biophys. Res. Commun. | pmid:3002349 |
| Peltier JB and Rossignol M | Auxin-induced differential sensitivity of the H+-ATPase in plasma membrane subfractions from tobacco cells. | 1996 | Biochem. Biophys. Res. Commun. | pmid:8605015 |
| Stan-Lotter H and Bragg PD | Electrophoretic determination of sulfhydryl groups and its application to complex protein samples, in vitro protein synthesis mixtures, and cross-linked proteins. | 1986 | Biochem. Cell Biol. | pmid:3755048 |
| Lima-Catelani AR et al. | Variation of genetic expression during development, revealed by esterase patterns in Aedes aegypti (Diptera, Culicidae). | 2004 | Biochem. Genet. | pmid:15168721 |
| Okochi VI et al. | Studies on the mechanism of adenosine transport in Trypanosoma vivax. | 1983 | Biochem. Int. | pmid:6679318 |
| Goswami A and Rosenberg IN | Reactive thiols in type II iodothyronine 5'-deiodinases: inactivation by iodoacetate. | 1989 | Biochem. Int. | pmid:2818601 |
| Berni R et al. | New crystalline derivatives of bovine liver rhodanese. | 1986 | Biochem. Int. | pmid:3460592 |
| Al-Saleh S et al. | On the disulphide bonds of rhodopsins. | 1987 | Biochem. J. | pmid:3675552 |
| Hawkins HC et al. | Redox properties and cross-linking of the dithiol/disulphide active sites of mammalian protein disulphide-isomerase. | 1991 | Biochem. J. | pmid:2025221 |
| Zawalich WS et al. | Metabolism and insulin-releasing capabilities of glucosamine and N-acetylglucosamine in isolated rat islets. | 1979 | Biochem. J. | pmid:384996 |
| Oteiza PI et al. | Cyst(e)ine residues of bovine white-matter proteolipid proteins. Role of disulphides in proteolipid conformation. | 1987 | Biochem. J. | pmid:3663175 |
| Nimmo GA and Nimmo HG | Studies of rat adipose-tissue microsomal glycerol phosphate acyltransferase. | 1984 | Biochem. J. | pmid:6508751 |
| Price NC and Stevens E | The refolding of denatured rabbit muscle creatine kinase. Search for intermediates in the refolding process and effect of modification at the reactive thiol group on refolding. | 1982 | Biochem. J. | pmid:6805463 |
| Cheng KW | Carboxymethylation of methionine residues in bovine pituitary luteinizing hormone and its subunits. Location of specifically modified methionine residues. | 1976 | Biochem. J. | pmid:999646 |
| Butler PE et al. | Characterization of meprin, a membrane-bound metalloendopeptidase from mouse kidney. | 1987 | Biochem. J. | pmid:3105525 |
| Allen G | Primary structure of the calcium ion-transporting adenosine triphosphatase of rabbit skeletal sarcoplasmic reticulum. Soluble peptides from the alpha-chymotryptic digest of the carboxymethylated protein. | 1980 | Biochem. J. | pmid:6234879 |
| Allen G et al. | The primary structure of the calcium ion-transporting adenosine triphosphatase protein of rabbit skeletal sarcoplasmic reticulum. Peptides derived from digestion with cyanogen bromide, and the sequences of three long extramembranous segments. | 1980 | Biochem. J. | pmid:6234881 |
| Buttle DJ et al. | Inhibition of interleukin 1-stimulated cartilage proteoglycan degradation by a lipophilic inactivator of cysteine endopeptidases. | 1992 | Biochem. J. | pmid:1731753 |
| Anastasi A et al. | Cystatin, a protein inhibitor of cysteine proteinases. Improved purification from egg white, characterization, and detection in chicken serum. | 1983 | Biochem. J. | pmid:6409085 |
| Gounaris AD and Slater EE | Cathepsin B from human renal cortex. | 1982 | Biochem. J. | pmid:7138503 |
| Salamino F et al. | Site-directed activation of calpain is promoted by a membrane-associated natural activator protein. | 1993 | Biochem. J. | pmid:8439288 |
| Thomas AP and Halestrap AP | Identification of the protein responsible for pyruvate transport into rat liver and heart mitochondria by specific labelling with [3H]N-phenylmaleimide. | 1981 | Biochem. J. | pmid:7316989 |