| MeSH term | MeSH ID | Detail |
|---|---|---|
| Cystitis | D003556 | 23 associated lipids |
| Parkinsonian Disorders | D020734 | 20 associated lipids |
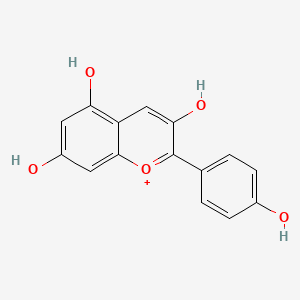
Pelargonidin
Pelargonidin is a lipid of Polyketides (PK) class. The involved functions are known as Uptake, Intestinal Absorption, glucose uptake, Process and Metabolic Inhibition. Pelargonidin often locates in Serosal, Mucous Membrane, brush border membrane, Membrane and Cell surface. The associated genes with Pelargonidin are SLC5A1 gene, SLC2A2 gene, Homologous Gene, F3 gene and CRSP3 gene. The related experimental models are Knock-out.
Cross Reference
Introduction
To understand associated biological information of Pelargonidin, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with Pelargonidin?
There are no associated biomedical information in the current reference collection.
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with Pelargonidin
PubChem Associated disorders and diseases
What pathways are associated with Pelargonidin
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with Pelargonidin?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with Pelargonidin?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with Pelargonidin?
There are no associated biomedical information in the current reference collection.
What genes are associated with Pelargonidin?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with Pelargonidin?
Knock-out
Knock-out are used in the study 'MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula.' (Zhao J et al., 2011).
Related references are published most in these journals:
| Model | Cross reference | Weighted score | Related literatures |
|---|
NCBI Entrez Crosslinks
All references with Pelargonidin
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Zhao J et al. | MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. | 2011 | Plant Cell | pmid:21467581 |
| Ono E et al. | Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). | 2010 | Plant Cell | pmid:20693356 |
| Hoshino A et al. | Spontaneous mutations of the flavonoid 3'-hydroxylase gene conferring reddish flowers in the three morning glory species. | 2003 | Plant Cell Physiol. | pmid:14581624 |
| Sato M et al. | Tissue culture-induced flower-color changes in Saintpaulia caused by excision of the transposon inserted in the flavonoid 3', 5' hydroxylase (F3'5'H) promoter. | 2011 | Plant Cell Rep. | pmid:21293860 |
| Nakatsuka T et al. | Production of red-flowered plants by genetic engineering of multiple flavonoid biosynthetic genes. | 2007 | Plant Cell Rep. | pmid:17639403 |
| Han Y et al. | Ectopic expression of apple F3'H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. | 2010 | Plant Physiol. | pmid:20357139 |
| Suzuki H et al. | cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme a:anthocyanidin 3-o-glucoside-6"-o-malonyltransferase from dahlia flowers. | 2002 | Plant Physiol. | pmid:12481098 |
| Xie DY et al. | Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. | 2004 | Plant Physiol. | pmid:14976232 |
| Dong X et al. | Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. | 2001 | Plant Physiol. | pmid:11553733 |
| Chiu LW et al. | The purple cauliflower arises from activation of a MYB transcription factor. | 2010 | Plant Physiol. | pmid:20855520 |