| MeSH term | MeSH ID | Detail |
|---|---|---|
| Inflammatory Bowel Diseases | D015212 | 9 associated lipids |
| Occupational Diseases | D009784 | 42 associated lipids |
| Metabolism, Inborn Errors | D008661 | 46 associated lipids |
| Amino Acid Metabolism, Inborn Errors | D000592 | 16 associated lipids |
| Olfaction Disorders | D000857 | 17 associated lipids |
| Maple Syrup Urine Disease | D008375 | 5 associated lipids |
| Acidosis, Renal Tubular | D000141 | 4 associated lipids |
| Fanconi Syndrome | D005198 | 4 associated lipids |
| Lipid Metabolism, Inborn Errors | D008052 | 26 associated lipids |
| Dwarfism | D004392 | 1 associated lipids |
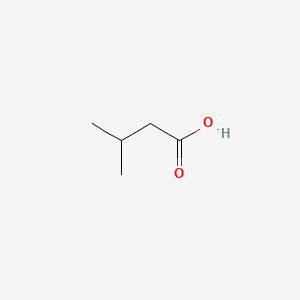
ISOVALERIC ACID
ISOVALERIC ACID is a lipid of Fatty Acyls (FA) class. Isovaleric acid is associated with abnormalities such as Infection, Lymphocele, Cyst, Abscess and Subgingival plaque. The involved functions are known as Biochemical Reaction, 5-(carboxyamino)imidazole ribonucleotide mutase activity, nitrate reductase activity, urease activity and colony morphology. Isovaleric acid often locates in Skeleton, Abdomen, Chromosomes, Tissue membrane and Microsomes. The associated genes with ISOVALERIC ACID are trypticase, Operon, KCNT1 gene, Genome and Reverse primer. The related lipids are Fatty Acids, Propionate, Fatty Acids, Unsaturated, Steroids and Promega. The related experimental models are Knock-out.
Cross Reference
Introduction
To understand associated biological information of ISOVALERIC ACID, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with ISOVALERIC ACID?
ISOVALERIC ACID is suspected in Exanthema, Isovaleryl-CoA dehydrogenase deficiency, Subgingival plaque, Dehydration, MAJOR DEPRESSIVE DISORDER 1, MAJOR DEPRESSIVE DISORDER 2 and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with ISOVALERIC ACID
PubChem Associated disorders and diseases
What pathways are associated with ISOVALERIC ACID
Lipid pathways are not clear in current pathway databases. We organized associated pathways with ISOVALERIC ACID through full-text articles, including metabolic pathways or pathways of biological mechanisms.
Related references are published most in these journals:
| Pathway name | Related literatures |
|---|
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with ISOVALERIC ACID?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
- Appl. Environ. Microbiol. (4)
- Int. J. Syst. Evol. Microbiol. (3)
- Microbiology (Reading, Engl.) (2)
- Others (10)
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with ISOVALERIC ACID?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with ISOVALERIC ACID?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with ISOVALERIC ACID?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with ISOVALERIC ACID?
Knock-out
Knock-out are used in the study '3-Hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase is involved in biosynthesis of isovaleryl-CoA in the myxobacterium Myxococcus xanthus during fruiting body formation.' (Bode HB et al., 2006) and Knock-out are used in the study 'Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized.' (Ruan Y et al., 2001).
Related references are published most in these journals:
| Model | Cross reference | Weighted score | Related literatures |
|---|
NCBI Entrez Crosslinks
All references with ISOVALERIC ACID
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Grégoire P et al. | Desulfosoma profundi sp. nov., a thermophilic sulfate-reducing bacterium isolated from a deep terrestrial geothermal spring in France. | 2012 | Antonie Van Leeuwenhoek | pmid:22120904 |
| Thierry A et al. | Conversion of L-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. | 2002 | Appl. Environ. Microbiol. | pmid:11823198 |
| Serrazanetti DI et al. | Acid stress-mediated metabolic shift in Lactobacillus sanfranciscensis LSCE1. | 2011 | Appl. Environ. Microbiol. | pmid:21335381 |
| Gogerty DS and Bobik TA | Formation of isobutene from 3-hydroxy-3-methylbutyrate by diphosphomevalonate decarboxylase. | 2010 | Appl. Environ. Microbiol. | pmid:20971863 |
| Aguilar JA et al. | The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa. | 2006 | Appl. Environ. Microbiol. | pmid:16517656 |
| Förster-Fromme K et al. | Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. | 2006 | Appl. Environ. Microbiol. | pmid:16820476 |
| Allison MJ et al. | Alternative pathways for biosynthesis of leucine and other amino acids in Bacteroides ruminicola and Bacteroides fragilis. | 1984 | Appl. Environ. Microbiol. | pmid:6440485 |
| Tanner RS and Wolfe RS | Nutritional requirements of Methanomicrobium mobile. | 1988 | Appl. Environ. Microbiol. | pmid:3377488 |
| Paczkowski S and Schütz S | Post-mortem volatiles of vertebrate tissue. | 2011 | Appl. Microbiol. Biotechnol. | pmid:21720824 |
| Thrash JC et al. | Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov.and Propionivibrio militaris, sp. nov. | 2010 | Appl. Microbiol. Biotechnol. | pmid:19921177 |