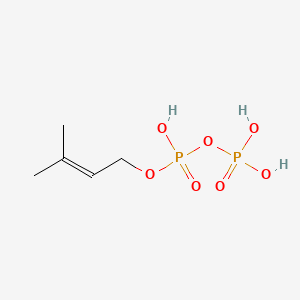
| Qian Q et al. |
Mechanistic studies on CymD: a tryptophan reverse N-prenyltransferase. |
2012 |
Biochemistry |
pmid:22935004
|
| Behr M et al. |
Remodeling of cytokinin metabolism at infection sites of Colletotrichum graminicola on maize leaves. |
2012 |
Mol. Plant Microbe Interact. |
pmid:22746825
|
| Testa CA and Johnson LJ |
A whole-cell phenotypic screening platform for identifying methylerythritol phosphate pathway-selective inhibitors as novel antibacterial agents. |
2012 |
Antimicrob. Agents Chemother. |
pmid:22777049
|
| Lu XM et al. |
Map-based cloning of zb7 encoding an IPP and DMAPP synthase in the MEP pathway of maize. |
2012 |
Mol Plant |
pmid:22498772
|
| Song AA et al. |
Functional expression of an orchid fragrance gene in Lactococcus lactis. |
2012 |
Int J Mol Sci |
pmid:22408409
|
| Kuzuyama T and Seto H |
Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. |
2012 |
Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. |
pmid:22450534
|
| Trowbridge AM et al. |
Contribution of various carbon sources toward isoprene biosynthesis in poplar leaves mediated by altered atmospheric CO2 concentrations. |
2012 |
PLoS ONE |
pmid:22384238
|
| Yang ZB et al. |
Physiological and molecular analysis of the interaction between aluminium toxicity and drought stress in common bean (Phaseolus vulgaris). |
2012 |
J. Exp. Bot. |
pmid:22371077
|
| Suzuki N et al. |
Construction and analysis of EST libraries of the trans-polyisoprene producing plant, Eucommia ulmoides Oliver. |
2012 |
Planta |
pmid:22729820
|
| Jonnalagadda V et al. |
Isopentenyl diphosphate isomerase catalyzed reactions in D2O: product release limits the rate of this sluggish enzyme-catalyzed reaction. |
2012 |
J. Am. Chem. Soc. |
pmid:22471428
|
| Fischer MJ et al. |
Identification of a lysine residue important for the catalytic activity of yeast farnesyl diphosphate synthase. |
2011 |
Protein J. |
pmid:21643844
|
| Heaps NA and Poulter CD |
Synthesis and evaluation of chlorinated substrate analogues for farnesyl diphosphate synthase. |
2011 |
J. Org. Chem. |
pmid:21344952
|
| Thabet I et al. |
The subcellular localization of periwinkle farnesyl diphosphate synthase provides insight into the role of peroxisome in isoprenoid biosynthesis. |
2011 |
J. Plant Physiol. |
pmid:21872968
|
| Heaps NA and Poulter CD |
Type-2 isopentenyl diphosphate isomerase: evidence for a stepwise mechanism. |
2011 |
J. Am. Chem. Soc. |
pmid:22047048
|
| Umeda T et al. |
Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum. |
2011 |
Sci Rep |
pmid:22355528
|
| Wen W and Yu R |
Artemisinin biosynthesis and its regulatory enzymes: Progress and perspective. |
2011 |
Pharmacogn Rev |
pmid:22279377
|
| Davey MS et al. |
Human neutrophil clearance of bacterial pathogens triggers anti-microbial γδ T cell responses in early infection. |
2011 |
PLoS Pathog. |
pmid:21589907
|
| Baumeister S et al. |
Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. |
2011 |
PLoS ONE |
pmid:21573242
|
| Meier S et al. |
A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. |
2011 |
BMC Syst Biol |
pmid:21595952
|
| Lemuth K et al. |
Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. |
2011 |
Microb. Cell Fact. |
pmid:21521516
|