| MeSH term | MeSH ID | Detail |
|---|---|---|
| Candidiasis, Invasive | D058365 | 2 associated lipids |
| Anisakiasis | D017129 | 2 associated lipids |
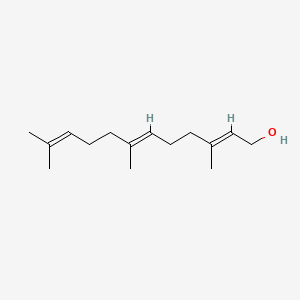
2E,6E-farnesol
2e,6e-farnesol is a lipid of Prenol Lipids (PR) class. 2e,6e-farnesol is associated with abnormalities such as Granulomatous Disease, Chronic, pathologic fistula and Cavitation. The involved functions are known as Regulation, Metabolic Inhibition, cholesterol biosynthetic process, Process and Transcription, Genetic. 2e,6e-farnesol often locates in Plasma membrane, Cytoplasmic matrix, cornified envelope, Epidermis and peroxisome. The associated genes with 2E,6E-farnesol are RAB3A gene, FOSL1 gene, CASP8AP2 gene, RCC1 gene and GALE gene. The related lipids are Sterols, Membrane Lipids and Steroids.
Cross Reference
Introduction
To understand associated biological information of 2E,6E-farnesol, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 2E,6E-farnesol?
2E,6E-farnesol is suspected in Granulomatous Disease, Chronic, pathologic fistula and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with 2E,6E-farnesol
PubChem Associated disorders and diseases
What pathways are associated with 2E,6E-farnesol
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 2E,6E-farnesol?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with 2E,6E-farnesol?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 2E,6E-farnesol?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with 2E,6E-farnesol?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with 2E,6E-farnesol?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with 2E,6E-farnesol
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Brehm-Stecher BF and Johnson EA | Sensitization of Staphylococcus aureus and Escherichia coli to antibiotics by the sesquiterpenoids nerolidol, farnesol, bisabolol, and apritone. | 2003 | Antimicrob. Agents Chemother. | pmid:14506058 |
| Pfister C et al. | Detection and quantification of farnesol-induced apoptosis in difficult primary cell cultures by TaqMan protein assay. | 2013 | Apoptosis | pmid:23315006 |
| Song L | A soluble form of phosphatase in Saccharomyces cerevisiae capable of converting farnesyl diphosphate into E,E-farnesol. | 2006 | Appl. Biochem. Biotechnol. | pmid:16484724 |
| Dionigi CP et al. | Effects of farnesol and the off-flavor derivative geosmin on Streptomyces tendae. | 1991 | Appl. Environ. Microbiol. | pmid:1785920 |
| Hornby JM et al. | Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. | 2001 | Appl. Environ. Microbiol. | pmid:11425711 |
| de Salas F et al. | Quorum-Sensing Mechanisms Mediated by Farnesol in Ophiostoma piceae: Effect on Secretion of Sterol Esterase. | 2015 | Appl. Environ. Microbiol. | pmid:25888179 |
| Ramage G et al. | Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. | 2002 | Appl. Environ. Microbiol. | pmid:12406738 |
| Hornby JM et al. | Inoculum size effect in dimorphic fungi: extracellular control of yeast-mycelium dimorphism in Ceratocystis ulmi. | 2004 | Appl. Environ. Microbiol. | pmid:15006753 |
| Mosel DD et al. | Farnesol concentrations required to block germ tube formation in Candida albicans in the presence and absence of serum. | 2005 | Appl. Environ. Microbiol. | pmid:16085901 |
| Nickerson KW et al. | Quorum sensing in dimorphic fungi: farnesol and beyond. | 2006 | Appl. Environ. Microbiol. | pmid:16751484 |
| Muramatsu M et al. | Accumulation of prenyl alcohols by terpenoid biosynthesis inhibitors in various microorganisms. | 2008 | Appl. Microbiol. Biotechnol. | pmid:18636253 |
| Brockmann H and Knobloch G | [A new bacteriochlorophyll from Rhodospirillum rubrum]. | 1972 | Arch Mikrobiol | pmid:4627398 |
| Suzue G et al. | Presence of squalene in Staphylococcus. | 1968 | Arch. Biochem. Biophys. | pmid:5650319 |
| Havel CM and Watson JA | Isopentenoid synthesis in isolated embryonic Drosophila cells: absolute, basal mevalonate synthesis rate determination. | 1992 | Arch. Biochem. Biophys. | pmid:1567218 |
| Dugan RE and Porter JW | Hog liver squalene synthetase: the partial purification of the particulate enzyme and kinetic analysis of the reaction. | 1972 | Arch. Biochem. Biophys. | pmid:4403691 |
| Jungalwala FB and Porter JW | Biosynthesis of phytoene from isopentenyl and farnesyl pyrophosphates by a partially purified tomato enzyme system. | 1967 | Arch. Biochem. Biophys. | pmid:4293186 |
| Meigs TE and Simoni RD | Farnesol as a regulator of HMG-CoA reductase degradation: characterization and role of farnesyl pyrophosphatase. | 1997 | Arch. Biochem. Biophys. | pmid:9281305 |
| Benedict CR et al. | Properties of farnesyl pyrophosphate synthetase of pig liver. | 1965 | Arch. Biochem. Biophys. | pmid:4284635 |
| Krishna G et al. | Enzymic conversion of farnesyl pyrophosphate to squalene. | 1966 | Arch. Biochem. Biophys. | pmid:4380977 |
| Xu L and Simoni RD | The inhibition of degradation of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase by sterol regulatory element binding protein cleavage-activating protein requires four phenylalanine residues in span 6 of HMG-CoA reductase transmembrane domain. | 2003 | Arch. Biochem. Biophys. | pmid:12781775 |