| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| pmid: | ||||
| Dickson RC et al. | Serine palmitoyltransferase. | 2000 | Meth. Enzymol. | pmid:10563304 |
| Gable K et al. | Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. | 2000 | J. Biol. Chem. | pmid:10713067 |
| Hanada K et al. | Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. | 2000 | J. Biol. Chem. | pmid:10722674 |
| Hanada K et al. | D-Serine inhibits serine palmitoyltransferase, the enzyme catalyzing the initial step of sphingolipid biosynthesis. | 2000 | FEBS Lett. | pmid:10828452 |
| Ardail D et al. | Occurrence of ceramides and neutral glycolipids with unusual long-chain base composition in purified rat liver mitochondria. | 2001 | FEBS Lett. | pmid:11163764 |
| Linn SC et al. | Regulation of de novo sphingolipid biosynthesis and the toxic consequences of its disruption. | 2001 | Biochem. Soc. Trans. | pmid:11709083 |
| Tamura K et al. | Characterization of an Arabidopsis cDNA encoding a subunit of serine palmitoyltransferase, the initial enzyme in sphingolipid biosynthesis. | 2001 | Plant Cell Physiol. | pmid:11726713 |
| Monaghan E et al. | Mutations in the Lcb2p subunit of serine palmitoyltransferase eliminate the requirement for the TSC3 gene in Saccharomyces cerevisiae. | 2002 | Yeast | pmid:12185836 |
| Verhoeven K et al. | SPTLC1 mutation in twin sisters with hereditary sensory neuropathy type I. | 2004 | Neurology | pmid:15037712 |
| Acharya U and Acharya JK | Enzymes of sphingolipid metabolism in Drosophila melanogaster. | 2005 | Cell. Mol. Life Sci. | pmid:15666085 |
| Merrill AH et al. | Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. | 2005 | Methods | pmid:15894491 |
| Yi H et al. | Direct evidence for the function of FUM13 in 3-ketoreduction of mycotoxin fumonisins in Fusarium verticillioides. | 2005 | J. Agric. Food Chem. | pmid:15969533 |
| Fornarotto M et al. | Sphingolipid biosynthesis in pathogenic fungi: identification and characterization of the 3-ketosphinganine reductase activity of Candida albicans and Aspergillus fumigatus. | 2006 | Biochim. Biophys. Acta | pmid:16431155 |
| Ikushiro H et al. | Molecular characterization of membrane-associated soluble serine palmitoyltransferases from Sphingobacterium multivorum and Bdellovibrio stolpii. | 2007 | J. Bacteriol. | pmid:17557831 |
| Pruett ST et al. | Biodiversity of sphingoid bases ("sphingosines") and related amino alcohols. | 2008 | J. Lipid Res. | pmid:18499644 |
| Gupta SD et al. | Tsc10p and FVT1: topologically distinct short-chain reductases required for long-chain base synthesis in yeast and mammals. | 2009 | J. Lipid Res. | pmid:19141869 |
| Song WQ et al. | Characterization of two cotton cDNAs encoding trans-2-enoyl-CoA reductase reveals a putative novel NADPH-binding motif. | 2009 | J. Exp. Bot. | pmid:19286916 |
| Ikushiro H et al. | Structural insights into the enzymatic mechanism of serine palmitoyltransferase from Sphingobacterium multivorum. | 2009 | J. Biochem. | pmid:19564159 |
| Seamen E et al. | P-type ATPase TAT-2 negatively regulates monomethyl branched-chain fatty acid mediated function in post-embryonic growth and development in C. elegans. | 2009 | PLoS Genet. | pmid:19662161 |
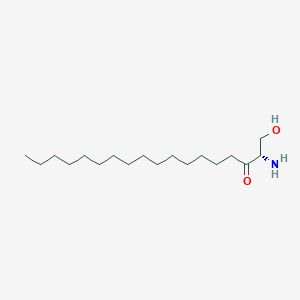
3-Ketosphinganine
3-Ketosphinganine is a lipid of Sphingolipids (SP) class. The involved functions are known as Anabolism and establishment and maintenance of localization. 3-ketosphinganine often locates in Membrane and membrane raft.
Cross Reference
Introduction
To understand associated biological information of 3-Ketosphinganine, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 3-Ketosphinganine?
There are no associated biomedical information in the current reference collection.
No disease MeSH terms mapped to the current reference collection.
PubChem Associated disorders and diseases
What pathways are associated with 3-Ketosphinganine
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 3-Ketosphinganine?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with 3-Ketosphinganine?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 3-Ketosphinganine?
There are no associated biomedical information in the current reference collection.
What genes are associated with 3-Ketosphinganine?
There are no associated biomedical information in the current reference collection.
What common seen animal models are associated with 3-Ketosphinganine?
There are no associated biomedical information in the current reference collection.