| MeSH term | MeSH ID | Detail |
|---|---|---|
| Peripheral Arterial Disease | D058729 | 7 associated lipids |
| Vascular System Injuries | D057772 | 2 associated lipids |
| Atherosclerosis | D050197 | 85 associated lipids |
| Stroke | D020521 | 32 associated lipids |
| Brain Infarction | D020520 | 17 associated lipids |
| Carotid Artery Injuries | D020212 | 8 associated lipids |
| Vasculitis | D014657 | 14 associated lipids |
| Thrombosis | D013927 | 49 associated lipids |
| Thromboangiitis Obliterans | D013919 | 4 associated lipids |
| Starvation | D013217 | 47 associated lipids |
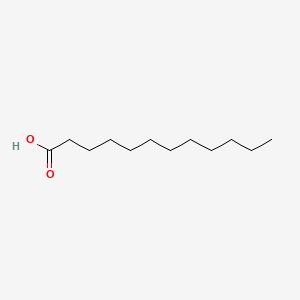
lauric acid
lauric acid is a lipid of Fatty Acyls (FA) class. Lauric acid is associated with abnormalities such as Infection, Renal tubular disorder, Hypertensive disease, Obesity and Mycoses. The involved functions are known as Transcription, Genetic, Signal Transduction, Mutation, metaplastic cell transformation and Anabolism. Lauric acid often locates in Skin, Plasma membrane, Cytoplasmic matrix, Body tissue and Palmar surface. The associated genes with lauric acid are Gene Family, SLC33A1 gene, Homologous Gene, Open Reading Frames and P4HTM gene. The related lipids are Fatty Acids, Oleic Acids, Palmitates, Stearates and 9,11-linoleic acid.
Cross Reference
Introduction
To understand associated biological information of lauric acid, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with lauric acid?
lauric acid is suspected in Renal tubular disorder, Hypertensive disease, Infection, Renal vascular disorder, Obesity, Mycoses and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with lauric acid
PubChem Associated disorders and diseases
What pathways are associated with lauric acid
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with lauric acid?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with lauric acid?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with lauric acid?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with lauric acid?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with lauric acid?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with lauric acid
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Bridgham JT et al. | Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. | 2010 | PLoS Biol. | pmid:20957188 |
| Chopra T et al. | Novel intermolecular iterative mechanism for biosynthesis of mycoketide catalyzed by a bimodular polyketide synthase. | 2008 | PLoS Biol. | pmid:18613748 |
| Hernández-Galicia E et al. | Monoglycerides and fatty acids from Ibervillea sonorae root: isolation and hypoglycemic activity. | 2007 | Planta Med. | pmid:17318782 |
| Rösler TW et al. | Analysis of the hydrodistillate from the fruits of Serenoa repens. | 2009 | Planta Med. | pmid:19140098 |
| Crane J et al. | Triacylglycerols determine the unusual storage physiology of Cuphea seed. | 2003 | Planta | pmid:12743824 |
| Hunzicker GM | A novel regulatory system in plants involving medium-chain fatty acids. | 2009 | Planta | pmid:19876644 |
| Wiberg E et al. | Fatty acid distribution and lipid metabolism in developing seeds of laurate-producing rape (Brassica napus L.). | 1997 | Planta | pmid:9431681 |
| Wiberg E et al. | The distribution of caprylate, caprate and laurate in lipids from developing and mature seeds of transgenic Brassica napus L. | 2000 | Planta | pmid:11219581 |
| Madey E et al. | Isolation and characterization of lipid in phloem sap of canola. | 2002 | Planta | pmid:11925046 |
| Lawo NC et al. | The volatile metabolome of grapevine roots: first insights into the metabolic response upon phylloxera attack. | 2011 | Plant Physiol. Biochem. | pmid:21764593 |
| Dobritsa AA et al. | CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. | 2009 | Plant Physiol. | pmid:19700560 |
| Quilichini TD et al. | ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. | 2010 | Plant Physiol. | pmid:20732973 |
| Joyard J and Stumpf PK | Synthesis of Long-Chain Acyl-CoA in Chloroplast Envelope Membranes. | 1981 | Plant Physiol. | pmid:16661656 |
| Benveniste I et al. | Cytochrome P-450-Dependent omega-Hydroxylation of Lauric Acid by Microsomes from Pea Seedlings. | 1982 | Plant Physiol. | pmid:16662431 |
| Cao YZ and Huang AH | Acyl coenzyme a preference of diacylglycerol acyltransferase from the maturing seeds of cuphea, maize, rapeseed, and canola. | 1987 | Plant Physiol. | pmid:16665518 |
| Saffert A et al. | A dual function alpha-dioxygenase-peroxidase and NAD(+) oxidoreductase active enzyme from germinating pea rationalizing alpha-oxidation of fatty acids in plants. | 2000 | Plant Physiol. | pmid:10938370 |
| Nguyen HT et al. | Metabolic engineering of seeds can achieve levels of omega-7 fatty acids comparable with the highest levels found in natural plant sources. | 2010 | Plant Physiol. | pmid:20943853 |
| Dussert S et al. | Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. | 2013 | Plant Physiol. | pmid:23735505 |
| Zhang D et al. | OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. | 2010 | Plant Physiol. | pmid:20610705 |
| Dobritsa AA et al. | LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. | 2010 | Plant Physiol. | pmid:20442277 |