| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Sastalla I et al. | Activation of the latent PlcR regulon in Bacillus anthracis. | 2010 | Microbiology (Reading, Engl.) | pmid:20688829 |
| Li X et al. | Reverse-genetic analysis of the two biotin-containing subunit genes of the heteromeric acetyl-coenzyme A carboxylase in Arabidopsis indicates a unidirectional functional redundancy. | 2011 | Plant Physiol. | pmid:21030508 |
| Tanabe Y et al. | Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. | 2011 | J. Biol. Chem. | pmid:21730067 |
| Bonde BK et al. | Differential producibility analysis (DPA) of transcriptomic data with metabolic networks: deconstructing the metabolic response of M. tuberculosis. | 2011 | PLoS Comput. Biol. | pmid:21738454 |
| Woong Park S et al. | Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. | 2011 | PLoS Pathog. | pmid:21980288 |
| Waller JC et al. | Mitochondrial and plastidial COG0354 proteins have folate-dependent functions in iron-sulphur cluster metabolism. | 2012 | J. Exp. Bot. | pmid:21984653 |
| Magliano P et al. | Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi. | 2011 | J. Biol. Chem. | pmid:21998305 |
| Charles H et al. | A genomic reappraisal of symbiotic function in the aphid/Buchnera symbiosis: reduced transporter sets and variable membrane organisations. | 2011 | PLoS ONE | pmid:22229056 |
| Nijkamp JF et al. | De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. | 2012 | Microb. Cell Fact. | pmid:22448915 |
| Cobessi D et al. | Biochemical and structural characterization of the Arabidopsis bifunctional enzyme dethiobiotin synthetase-diaminopelargonic acid aminotransferase: evidence for substrate channeling in biotin synthesis. | 2012 | Plant Cell | pmid:22547782 |
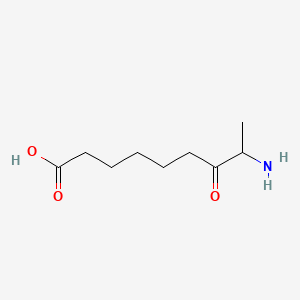
8-amino-7-oxononanoic acid
8-amino-7-oxononanoic acid is a lipid of Fatty Acyls (FA) class. The involved functions are known as seed development.
Cross Reference
Introduction
To understand associated biological information of 8-amino-7-oxononanoic acid, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 8-amino-7-oxononanoic acid?
There are no associated biomedical information in the current reference collection.
No disease MeSH terms mapped to the current reference collection.
PubChem Associated disorders and diseases
What pathways are associated with 8-amino-7-oxononanoic acid
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 8-amino-7-oxononanoic acid?
There are no associated biomedical information in the current reference collection.
What functions are associated with 8-amino-7-oxononanoic acid?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 8-amino-7-oxononanoic acid?
There are no associated biomedical information in the current reference collection.
What genes are associated with 8-amino-7-oxononanoic acid?
There are no associated biomedical information in the current reference collection.
What common seen animal models are associated with 8-amino-7-oxononanoic acid?
There are no associated biomedical information in the current reference collection.