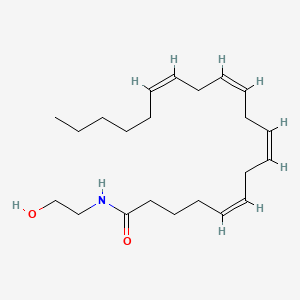
Anandamide
Anandamide is a lipid of Fatty Acyls (FA) class. Anandamide is associated with abnormalities such as Dehydration. The involved functions are known as Process, Phenomenon, Phosphorylation, Catabolic Process and Gene Expression. Anandamide often locates in Nuchal region, Microglial and Hepatic. The associated genes with Anandamide are SGPL1 gene, SPTLC1 gene, RPSA gene, KDSR gene and SMPD1 gene. The related lipids are Sphingolipids, Lipopolysaccharides, Lysophospholipids, LYSO-PC and lysophosphatidylethanolamine.
Cross Reference
Introduction
To understand associated biological information of Anandamide, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with Anandamide?
Anandamide is suspected in Dehydration and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with Anandamide
PubChem Associated disorders and diseases
What pathways are associated with Anandamide
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with Anandamide?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with Anandamide?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with Anandamide?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with Anandamide?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with Anandamide?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with Anandamide
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Czifra G et al. | Endocannabinoids regulate growth and survival of human eccrine sweat gland-derived epithelial cells. | 2012 | J. Invest. Dermatol. | pmid:22513781 |
| Turcotte C et al. | Regulation of inflammation by cannabinoids, the endocannabinoids 2-arachidonoyl-glycerol and arachidonoyl-ethanolamide, and their metabolites. | 2015 | J. Leukoc. Biol. | pmid:25877930 |
| Oka S et al. | 2-arachidonoylglycerol, an endogenous cannabinoid receptor ligand, induces the migration of EoL-1 human eosinophilic leukemia cells and human peripheral blood eosinophils. | 2004 | J. Leukoc. Biol. | pmid:15316028 |
| Jackson AR et al. | Characterization of endocannabinoid-mediated induction of myeloid-derived suppressor cells involving mast cells and MCP-1. | 2014 | J. Leukoc. Biol. | pmid:24319288 |
| Glass M et al. | Misidentification of prostamides as prostaglandins. | 2005 | J. Lipid Res. | pmid:15863842 |
| Newberry EP et al. | Decreased body weight and hepatic steatosis with altered fatty acid ethanolamide metabolism in aged L-Fabp -/- mice. | 2012 | J. Lipid Res. | pmid:22327204 |
| Ambrosi S et al. | On the importance of anandamide structural features for its interactions with DPPC bilayers: effects on PLA2 activity. | 2005 | J. Lipid Res. | pmid:15961786 |
| Bojesen IN and Hansen HS | Membrane transport of anandamide through resealed human red blood cell membranes. | 2005 | J. Lipid Res. | pmid:15930521 |
| Weber A et al. | Formation of prostamides from anandamide in FAAH knockout mice analyzed by HPLC with tandem mass spectrometry. | 2004 | J. Lipid Res. | pmid:14729864 |
| Kantae V et al. | Quantitative profiling of endocannabinoids and related -acylethanolamines in human CSF using nano LC-MS/MS. | 2017 | J. Lipid Res. | pmid:27999147 |
| Long JZ et al. | An anatomical and temporal portrait of physiological substrates for fatty acid amide hydrolase. | 2011 | J. Lipid Res. | pmid:21097653 |
| Oddi S et al. | Pitfalls and solutions in assaying anandamide transport in cells. | 2010 | J. Lipid Res. | pmid:20447929 |
| Fezza F et al. | Characterization of biotin-anandamide, a novel tool for the visualization of anandamide accumulation. | 2008 | J. Lipid Res. | pmid:18316795 |
| Bojesen IN and Hansen HS | Binding of anandamide to bovine serum albumin. | 2003 | J. Lipid Res. | pmid:12837852 |
| Jacome-Sosa M et al. | Vaccenic acid suppresses intestinal inflammation by increasing anandamide and related N-acylethanolamines in the JCR:LA-cp rat. | 2016 | J. Lipid Res. | pmid:26891736 |
| Yang W et al. | Enzymatic formation of prostamide F2alpha from anandamide involves a newly identified intermediate metabolite, prostamide H2. | 2005 | J. Lipid Res. | pmid:16150817 |
| Patel S et al. | The postmortal accumulation of brain N-arachidonylethanolamine (anandamide) is dependent upon fatty acid amide hydrolase activity. | 2005 | J. Lipid Res. | pmid:15576840 |
| Bojesen IN and Hansen HS | Effect of an unstirred layer on the membrane permeability of anandamide. | 2006 | J. Lipid Res. | pmid:16365480 |
| Wollank Y et al. | Inhibition of FAAH confers increased stem cell migration via PPARα. | 2015 | J. Lipid Res. | pmid:26263913 |
| Clugston RD et al. | Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. | 2011 | J. Lipid Res. | pmid:21856784 |
| Markey SP et al. | Base- and acid-catalyzed interconversions of O-acyl- and N-acyl-ethanolamines: a cautionary note for lipid analyses. | 2000 | J. Lipid Res. | pmid:10744787 |
| Hillard CJ and Campbell WB | Biochemistry and pharmacology of arachidonylethanolamide, a putative endogenous cannabinoid. | 1997 | J. Lipid Res. | pmid:9458263 |
| Astarita G et al. | Identification of biosynthetic precursors for the endocannabinoid anandamide in the rat brain. | 2008 | J. Lipid Res. | pmid:17957091 |
| De Petrocellis L et al. | Chemical synthesis, pharmacological characterization, and possible formation in unicellular fungi of 3-hydroxy-anandamide. | 2009 | J. Lipid Res. | pmid:19017617 |
| McAllister SD et al. | An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. | 2003 | J. Med. Chem. | pmid:14613317 |
| Lambert DM and Fowler CJ | The endocannabinoid system: drug targets, lead compounds, and potential therapeutic applications. | 2005 | J. Med. Chem. | pmid:16078824 |
| Vandevoorde S et al. | Esters, retroesters, and a retroamide of palmitic acid: pool for the first selective inhibitors of N-palmitoylethanolamine-selective acid amidase. | 2003 | J. Med. Chem. | pmid:14521402 |
| Ng EW et al. | Unique analogues of anandamide: arachidonyl ethers and carbamates and norarachidonyl carbamates and ureas. | 1999 | J. Med. Chem. | pmid:10354405 |
| Schiano Moriello A et al. | Development of the first potential covalent inhibitors of anandamide cellular uptake. | 2006 | J. Med. Chem. | pmid:16570928 |
| Appendino G et al. | Oxyhomologues of anandamide and related endolipids: chemoselective synthesis and biological activity. | 2006 | J. Med. Chem. | pmid:16570929 |
| Tarzia G et al. | Design, synthesis, and structure-activity relationships of alkylcarbamic acid aryl esters, a new class of fatty acid amide hydrolase inhibitors. | 2003 | J. Med. Chem. | pmid:12773040 |
| López-RodrÃguez ML et al. | Design, synthesis, and biological evaluation of new inhibitors of the endocannabinoid uptake: comparison with effects on fatty acid amidohydrolase. | 2003 | J. Med. Chem. | pmid:12672252 |
| Tuccinardi T et al. | Cannabinoid CB2/CB1 selectivity. Receptor modeling and automated docking analysis. | 2006 | J. Med. Chem. | pmid:16451064 |
| Balas L et al. | Total synthesis of photoactivatable or fluorescent anandamide probes: novel bioactive compounds with angiogenic activity. | 2009 | J. Med. Chem. | pmid:19161308 |
| Hanus L et al. | Two new unsaturated fatty acid ethanolamides in brain that bind to the cannabinoid receptor. | 1993 | J. Med. Chem. | pmid:8411021 |
| Cisneros JA et al. | Structure-activity relationship of a series of inhibitors of monoacylglycerol hydrolysis--comparison with effects upon fatty acid amide hydrolase. | 2007 | J. Med. Chem. | pmid:17764163 |
| Barnett-Norris J et al. | Exploration of biologically relevant conformations of anandamide, 2-arachidonylglycerol, and their analogues using conformational memories. | 1998 | J. Med. Chem. | pmid:9822555 |
| MartÃn-Couce L et al. | Development of endocannabinoid-based chemical probes for the study of cannabinoid receptors. | 2011 | J. Med. Chem. | pmid:21675776 |
| Romero FA et al. | Potent and selective alpha-ketoheterocycle-based inhibitors of the anandamide and oleamide catabolizing enzyme, fatty acid amide hydrolase. | 2007 | J. Med. Chem. | pmid:17279740 |
| Salo OM et al. | 3D-QSAR studies on cannabinoid CB1 receptor agonists: G-protein activation as biological data. | 2006 | J. Med. Chem. | pmid:16420041 |
| Avraham Y et al. | Novel acylethanolamide derivatives that modulate body weight through enhancement of hypothalamic pro-opiomelanocortin (POMC) and/or decreased neuropeptide Y (NPY). | 2013 | J. Med. Chem. | pmid:23384387 |
| Onnis V et al. | Synthesis and evaluation of paracetamol esters as novel fatty acid amide hydrolase inhibitors. | 2010 | J. Med. Chem. | pmid:20143779 |
| Li C et al. | High affinity electrophilic and photoactivatable covalent endocannabinoid probes for the CB1 receptor. | 2005 | J. Med. Chem. | pmid:16190768 |
| van der Stelt M et al. | Oxygenated metabolites of anandamide and 2-arachidonoylglycerol: conformational analysis and interaction with cannabinoid receptors, membrane transporter, and fatty acid amide hydrolase. | 2002 | J. Med. Chem. | pmid:12166944 |
| López-RodrÃguez ML et al. | Design, synthesis and biological evaluation of novel arachidonic acid derivatives as highly potent and selective endocannabinoid transporter inhibitors. | 2001 | J. Med. Chem. | pmid:11741470 |
| Seltzman HH et al. | Synthesis and pharmacological comparison of dimethylheptyl and pentyl analogs of anandamide. | 1997 | J. Med. Chem. | pmid:9357529 |
| Lin S et al. | Novel analogues of arachidonylethanolamide (anandamide): affinities for the CB1 and CB2 cannabinoid receptors and metabolic stability. | 1998 | J. Med. Chem. | pmid:9876105 |
| Fichera M et al. | A 3D-QSAR study on the structural requirements for binding to CB(1) and CB(2) cannabinoid receptors. | 2000 | J. Med. Chem. | pmid:10882356 |
| Brizzi A et al. | Resorcinol-sn-glycerol derivatives: novel 2-arachidonoylglycerol mimetics endowed with high affinity and selectivity for cannabinoid type 1 receptor. | 2011 | J. Med. Chem. | pmid:22044209 |
| Ortar G et al. | New N-arachidonoylserotonin analogues with potential "dual" mechanism of action against pain. | 2007 | J. Med. Chem. | pmid:18027904 |
| Ryan WJ et al. | Potent anandamide analogs: the effect of changing the length and branching of the end pentyl chain. | 1997 | J. Med. Chem. | pmid:9357528 |
| Appendino G et al. | Conformationally constrained fatty acid ethanolamides as cannabinoid and vanilloid receptor probes. | 2009 | J. Med. Chem. | pmid:19361197 |
| Thomas BF et al. | Structure-activity analysis of anandamide analogs: relationship to a cannabinoid pharmacophore. | 1996 | J. Med. Chem. | pmid:8558515 |
| Brizzi A et al. | New resorcinol-anandamide "hybrids" as potent cannabinoid receptor ligands endowed with antinociceptive activity in vivo. | 2009 | J. Med. Chem. | pmid:19331413 |
| Sheskin T et al. | Structural requirements for binding of anandamide-type compounds to the brain cannabinoid receptor. | 1997 | J. Med. Chem. | pmid:9057852 |
| Feledziak M et al. | SAR and LC/MS studies of β-lactamic inhibitors of human fatty acid amide hydrolase (hFAAH): evidence of a nonhydrolytic process. | 2011 | J. Med. Chem. | pmid:21899370 |
| Lang W et al. | Substrate specificity and stereoselectivity of rat brain microsomal anandamide amidohydrolase. | 1999 | J. Med. Chem. | pmid:10072686 |
| Brizzi A et al. | Design, synthesis, and binding studies of new potent ligands of cannabinoid receptors. | 2005 | J. Med. Chem. | pmid:16279794 |
| Khanolkar AD et al. | Head group analogs of arachidonylethanolamide, the endogenous cannabinoid ligand. | 1996 | J. Med. Chem. | pmid:8893848 |
| Palermo G et al. | Covalent inhibitors of fatty acid amide hydrolase: a rationale for the activity of piperidine and piperazine aryl ureas. | 2011 | J. Med. Chem. | pmid:21830831 |
| Barnett-Norris J et al. | Conformational memories and the endocannabinoid binding site at the cannabinoid CB1 receptor. | 2002 | J. Med. Chem. | pmid:12166938 |
| Wyffels L et al. | Synthesis, in vitro and in vivo evaluation, and radiolabeling of aryl anandamide analogues as candidate radioligands for in vivo imaging of fatty acid amide hydrolase in the brain. | 2009 | J. Med. Chem. | pmid:19719235 |
| Tong W et al. | Derivation of a pharmacophore model for anandamide using constrained conformational searching and comparative molecular field analysis. | 1998 | J. Med. Chem. | pmid:9784095 |
| Vandevoorde S et al. | Modifications of the ethanolamine head in N-palmitoylethanolamine: synthesis and evaluation of new agents interfering with the metabolism of anandamide. | 2003 | J. Med. Chem. | pmid:12672243 |
| Lynch DL and Reggio PH | Molecular dynamics simulations of the endocannabinoid N-arachidonoylethanolamine (anandamide) in a phospholipid bilayer: probing structure and dynamics. | 2005 | J. Med. Chem. | pmid:16033262 |
| Ueda N et al. | A hydrolase enzyme inactivating endogenous ligands for cannabinoid receptors. | 1998 | J. Med. Invest. | pmid:9864962 |
| Amorós I et al. | Endocannabinoids and cannabinoid analogues block human cardiac Kv4.3 channels in a receptor-independent manner. | 2010 | J. Mol. Cell. Cardiol. | pmid:19616555 |
| Weis F et al. | Substantially altered expression pattern of cannabinoid receptor 2 and activated endocannabinoid system in patients with severe heart failure. | 2010 | J. Mol. Cell. Cardiol. | pmid:19931541 |
| Hoyer FF et al. | Inhibition of endocannabinoid-degrading enzyme fatty acid amide hydrolase increases atherosclerotic plaque vulnerability in mice. | 2014 | J. Mol. Cell. Cardiol. | pmid:24286707 |
| Currie S et al. | IP(3)R-mediated Ca(2+) release is modulated by anandamide in isolated cardiac nuclei. | 2008 | J. Mol. Cell. Cardiol. | pmid:18692061 |
| D'Argenio G et al. | Overactivity of the intestinal endocannabinoid system in celiac disease and in methotrexate-treated rats. | 2007 | J. Mol. Med. | pmid:17396241 |
| Iuvone T et al. | Cannabinoid CB1 receptor stimulation affords neuroprotection in MPTP-induced neurotoxicity by attenuating S100B up-regulation in vitro. | 2007 | J. Mol. Med. | pmid:17639288 |
| Catani MV et al. | Expression of the endocannabinoid system in the bi-potential HEL cell line: commitment to the megakaryoblastic lineage by 2-arachidonoylglycerol. | 2009 | J. Mol. Med. | pmid:18820887 |
| Wang ZY et al. | Attenuation of cystitis and pain sensation in mice lacking fatty acid amide hydrolase. | 2015 | J. Mol. Neurosci. | pmid:25374388 |
| Lagalwar S et al. | Anandamides inhibit binding to the muscarinic acetylcholine receptor. | 1999 Aug-Oct | J. Mol. Neurosci. | pmid:10691292 |
| Naidoo V et al. | A new generation fatty acid amide hydrolase inhibitor protects against kainate-induced excitotoxicity. | 2011 | J. Mol. Neurosci. | pmid:21069475 |
| Hillard CJ et al. | Studies of anandamide accumulation inhibitors in cerebellar granule neurons: comparison to inhibition of fatty acid amide hydrolase. | 2007 | J. Mol. Neurosci. | pmid:17901541 |
| Han B et al. | Semiplenamides A-G, fatty acid amides from a Papua New Guinea collection of the marine cyanobacterium Lyngbya semiplena. | 2003 | J. Nat. Prod. | pmid:14575438 |
| Raduner S et al. | Self-assembling cannabinomimetics: supramolecular structures of N-alkyl amides. | 2007 | J. Nat. Prod. | pmid:17497806 |
| Hillard CJ et al. | Characterization of ligand binding to the cannabinoid receptor of rat brain membranes using a novel method: application to anandamide. | 1995 | J. Neurochem. | pmid:7830060 |
| Lam PM et al. | Activation of recombinant human TRPV1 receptors expressed in SH-SY5Y human neuroblastoma cells increases [Ca(2+)](i), initiates neurotransmitter release and promotes delayed cell death. | 2007 | J. Neurochem. | pmid:17442052 |
| Subbanna S et al. | Postnatal ethanol exposure alters levels of 2-arachidonylglycerol-metabolizing enzymes and pharmacological inhibition of monoacylglycerol lipase does not cause neurodegeneration in neonatal mice. | 2015 | J. Neurochem. | pmid:25857698 |
| Pearlman RJ et al. | Arachidonic acid and anandamide have opposite modulatory actions at the glycine transporter, GLYT1a. | 2003 | J. Neurochem. | pmid:12558979 |
| Maccarrone M et al. | Enhanced anandamide degradation is associated with neuronal apoptosis induced by the HIV-1 coat glycoprotein gp120 in the rat neocortex. | 2004 | J. Neurochem. | pmid:15147522 |
| Maccarrone M et al. | Levodopa treatment reverses endocannabinoid system abnormalities in experimental parkinsonism. | 2003 | J. Neurochem. | pmid:12716433 |
| Ahluwalia J et al. | Activation of capsaicin-sensitive primary sensory neurones induces anandamide production and release. | 2003 | J. Neurochem. | pmid:12558978 |
| De Lago E et al. | Acyl-based anandamide uptake inhibitors cause rapid toxicity to C6 glioma cells at pharmacologically relevant concentrations. | 2006 | J. Neurochem. | pmid:16899063 |
| Sarmad S et al. | Depolarizing and calcium-mobilizing stimuli fail to enhance synthesis and release of endocannabinoids from rat brain cerebral cortex slices. | 2011 | J. Neurochem. | pmid:21375532 |
| Hansen HH et al. | Accumulation of the anandamide precursor and other N-acylethanolamine phospholipids in infant rat models of in vivo necrotic and apoptotic neuronal death. | 2001 | J. Neurochem. | pmid:11145976 |
| Nogueron MI et al. | Cannabinoid receptor agonists inhibit depolarization-induced calcium influx in cerebellar granule neurons. | 2001 | J. Neurochem. | pmid:11677265 |
| Vogel Z et al. | Anandamide, a brain endogenous compound, interacts specifically with cannabinoid receptors and inhibits adenylate cyclase. | 1993 | J. Neurochem. | pmid:8515284 |
| Jarrahian A et al. | Structure-activity relationships among N-arachidonylethanolamine (Anandamide) head group analogues for the anandamide transporter. | 2000 | J. Neurochem. | pmid:10820223 |
| Di Marzo V et al. | Enhancement of anandamide formation in the limbic forebrain and reduction of endocannabinoid contents in the striatum of delta9-tetrahydrocannabinol-tolerant rats. | 2000 | J. Neurochem. | pmid:10737621 |
| Di Marzo V et al. | Levels, metabolism, and pharmacological activity of anandamide in CB(1) cannabinoid receptor knockout mice: evidence for non-CB(1), non-CB(2) receptor-mediated actions of anandamide in mouse brain. | 2000 | J. Neurochem. | pmid:11080195 |
| Rubino T et al. | Loss of cannabinoid-stimulated guanosine 5'-O-(3-[(35)S]Thiotriphosphate) binding without receptor down-regulation in brain regions of anandamide-tolerant rats. | 2000 | J. Neurochem. | pmid:11080200 |
| Wiles AL et al. | N-Arachidonyl-glycine inhibits the glycine transporter, GLYT2a. | 2006 | J. Neurochem. | pmid:16899062 |
| Cadogan AK et al. | Influence of cannabinoids on electrically evoked dopamine release and cyclic AMP generation in the rat striatum. | 1997 | J. Neurochem. | pmid:9282935 |
| Hill MN et al. | Regional alterations in the endocannabinoid system in an animal model of depression: effects of concurrent antidepressant treatment. | 2008 | J. Neurochem. | pmid:18643796 |
| Placzek EA et al. | Mechanisms for recycling and biosynthesis of endogenous cannabinoids anandamide and 2-arachidonylglycerol. | 2008 | J. Neurochem. | pmid:18778304 |
| Martin GG et al. | FABP-1 gene ablation impacts brain endocannabinoid system in male mice. | 2016 | J. Neurochem. | pmid:27167970 |