| MeSH term | MeSH ID | Detail |
|---|---|---|
| Hemolysis | D006461 | 131 associated lipids |
| Colonic Neoplasms | D003110 | 161 associated lipids |
| Alcoholism | D000437 | 27 associated lipids |
| Neoplasms, Glandular and Epithelial | D009375 | 12 associated lipids |
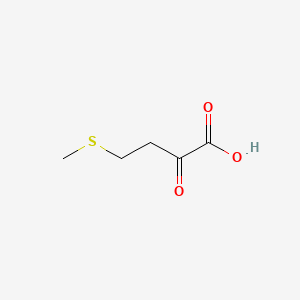
4-methylthio-2-oxobutanoic acid
4-methylthio-2-oxobutanoic acid is a lipid of Fatty Acyls (FA) class. The involved functions are known as polyamine biosynthetic process, Anabolism, enzyme activity, enzyme pathway and Process. The associated genes with 4-methylthio-2-oxobutanoic acid are 2-keto-4-methylthiobutyric acid, MTAP gene, Homologous Gene, 2-keto-4-thiomethylbutyrate and Alleles. The related lipids are alpha-ketocaproic acid.
Cross Reference
Introduction
To understand associated biological information of 4-methylthio-2-oxobutanoic acid, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with 4-methylthio-2-oxobutanoic acid?
There are no associated biomedical information in the current reference collection.
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with 4-methylthio-2-oxobutanoic acid
PubChem Associated disorders and diseases
What pathways are associated with 4-methylthio-2-oxobutanoic acid
There are no associated biomedical information in the current reference collection.
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with 4-methylthio-2-oxobutanoic acid?
There are no associated biomedical information in the current reference collection.
What functions are associated with 4-methylthio-2-oxobutanoic acid?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with 4-methylthio-2-oxobutanoic acid?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with 4-methylthio-2-oxobutanoic acid?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with 4-methylthio-2-oxobutanoic acid?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with 4-methylthio-2-oxobutanoic acid
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Splivallo R et al. | Truffles regulate plant root morphogenesis via the production of auxin and ethylene. | 2009 | Plant Physiol. | pmid:19535471 |
| Kabuyama Y et al. | A mediator of Rho-dependent invasion moonlights as a methionine salvage enzyme. | 2009 | Mol. Cell Proteomics | pmid:19620624 |
| Fun HK et al. | Ethyl 2-[(4-chloro-phen-yl)hydrazono]-3-oxobutanoate. | 2009 | Acta Crystallogr Sect E Struct Rep Online | pmid:21583848 |
| Knill T et al. | Arabidopsis thaliana encodes a bacterial-type heterodimeric isopropylmalate isomerase involved in both Leu biosynthesis and the Met chain elongation pathway of glucosinolate formation. | 2009 | Plant Mol. Biol. | pmid:19597944 |
| GarcÃa-GarcÃa M et al. | Production of the apoptotic cellular mediator 4-Methylthio-2-oxobutyric acid by using an enzymatic stirred tank reactor with in situ product removal. | 2008 Jan-Feb | Biotechnol. Prog. | pmid:18092800 |
| Hernández-Montes G et al. | The hidden universal distribution of amino acid biosynthetic networks: a genomic perspective on their origins and evolution. | 2008 | Genome Biol. | pmid:18541022 |
| Pirkov I et al. | A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. | 2008 | FEBS J. | pmid:18625006 |
| Chavali AK et al. | Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. | 2008 | Mol. Syst. Biol. | pmid:18364711 |
| Chai SC et al. | Characterization of metal binding in the active sites of acireductone dioxygenase isoforms from Klebsiella ATCC 8724. | 2008 | Biochemistry | pmid:18237192 |
| Knill T et al. | Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. | 2008 | Plant Physiol. | pmid:18162591 |