| MeSH term | MeSH ID | Detail |
|---|---|---|
| Melanoma | D008545 | 69 associated lipids |
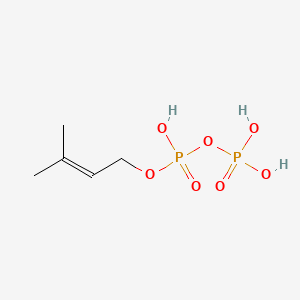
Dimethylallyl pyrophosphate
Dimethylallyl pyrophosphate is a lipid of Prenol Lipids (PR) class. Dimethylallyl pyrophosphate is associated with abnormalities such as Consumption-archaic term for TB and Wiskott-Aldrich Syndrome. The involved functions are known as Anabolism, Biochemical Pathway, Oxidation, Process and Chelating Activity [MoA]. Dimethylallyl pyrophosphate often locates in Chloroplasts, Plastids, chloroplast stroma, Cytosol and Cell membrane. The associated genes with Dimethylallyl pyrophosphate are IRF6 wt Allele and ADRBK1 gene. The related lipids are Sterols.
Cross Reference
Introduction
To understand associated biological information of Dimethylallyl pyrophosphate, we collected biological information of abnormalities, associated pathways, cellular/molecular locations, biological functions, related genes/proteins, lipids and common seen animal/experimental models with organized paragraphs from literatures.
What diseases are associated with Dimethylallyl pyrophosphate?
Dimethylallyl pyrophosphate is suspected in and other diseases in descending order of the highest number of associated sentences.
Related references are mostly published in these journals:
| Disease | Cross reference | Weighted score | Related literature |
|---|
Possible diseases from mapped MeSH terms on references
We collected disease MeSH terms mapped to the references associated with Dimethylallyl pyrophosphate
PubChem Associated disorders and diseases
What pathways are associated with Dimethylallyl pyrophosphate
Lipid pathways are not clear in current pathway databases. We organized associated pathways with Dimethylallyl pyrophosphate through full-text articles, including metabolic pathways or pathways of biological mechanisms.
Related references are published most in these journals:
| Pathway name | Related literatures |
|---|
PubChem Biomolecular Interactions and Pathways
Link to PubChem Biomolecular Interactions and PathwaysWhat cellular locations are associated with Dimethylallyl pyrophosphate?
Visualization in cellular structure
Associated locations are in red color. Not associated locations are in black.
Related references are published most in these journals:
| Location | Cross reference | Weighted score | Related literatures |
|---|
What functions are associated with Dimethylallyl pyrophosphate?
Related references are published most in these journals:
| Function | Cross reference | Weighted score | Related literatures |
|---|
What lipids are associated with Dimethylallyl pyrophosphate?
Related references are published most in these journals:
| Lipid concept | Cross reference | Weighted score | Related literatures |
|---|
What genes are associated with Dimethylallyl pyrophosphate?
Related references are published most in these journals:
| Gene | Cross reference | Weighted score | Related literatures |
|---|
What common seen animal models are associated with Dimethylallyl pyrophosphate?
There are no associated biomedical information in the current reference collection.
NCBI Entrez Crosslinks
All references with Dimethylallyl pyrophosphate
Download all related citations| Authors | Title | Published | Journal | PubMed Link |
|---|---|---|---|---|
| Metzger U et al. | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. | 2009 | Proc. Natl. Acad. Sci. U.S.A. | pmid:19706516 |
| Xu Q et al. | Comparative transcripts profiling reveals new insight into molecular processes regulating lycopene accumulation in a sweet orange (Citrus sinensis) red-flesh mutant. | 2009 | BMC Genomics | pmid:19922663 |
| Chang PK et al. | Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae. | 2009 | Toxins (Basel) | pmid:22069533 |
| Wang W et al. | Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. | 2009 | BMC Genomics | pmid:19818120 |
| de Souza W and Rodrigues JC | Sterol Biosynthesis Pathway as Target for Anti-trypanosomatid Drugs. | 2009 | Interdiscip Perspect Infect Dis | pmid:19680554 |
| Zhang J et al. | AtSOFL1 and AtSOFL2 act redundantly as positive modulators of the endogenous content of specific cytokinins in Arabidopsis. | 2009 | PLoS ONE | pmid:20011053 |
| Rotter A et al. | Gene expression profiling in susceptible interaction of grapevine with its fungal pathogen Eutypa lata: extending MapMan ontology for grapevine. | 2009 | BMC Plant Biol. | pmid:19656401 |
| Vandermoten S et al. | New insights into short-chain prenyltransferases: structural features, evolutionary history and potential for selective inhibition. | 2009 | Cell. Mol. Life Sci. | pmid:19633972 |
| Chung JA et al. | Purification of prenylated proteins by affinity chromatography on cyclodextrin-modified agarose. | 2009 | Anal. Biochem. | pmid:18834849 |
| Alagna F et al. | Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development. | 2009 | BMC Genomics | pmid:19709400 |